Phases. Today is Rare Disease Day. I would like to use this opportunity to explain some of the phenotype science that is critical for rare diseases. In contrast to common disorders, rare diseases face an unusual challenge. Once identified, the overall rareness of these condition poses the question of where phenotypes begin and where they end. For rare genetic disorders, is the phenotype of the first individual identified with a rare disease characteristic, or is there a larger spectrum that we should be aware of? Enter the various approaches to phenotype science that aim to decipher the full depth of clinical features associated with rare diseases. In order to understand the various approaches to rare diseases phenotypes, I would like to suggest a somewhat unusual analogy: phenotypes are like water.
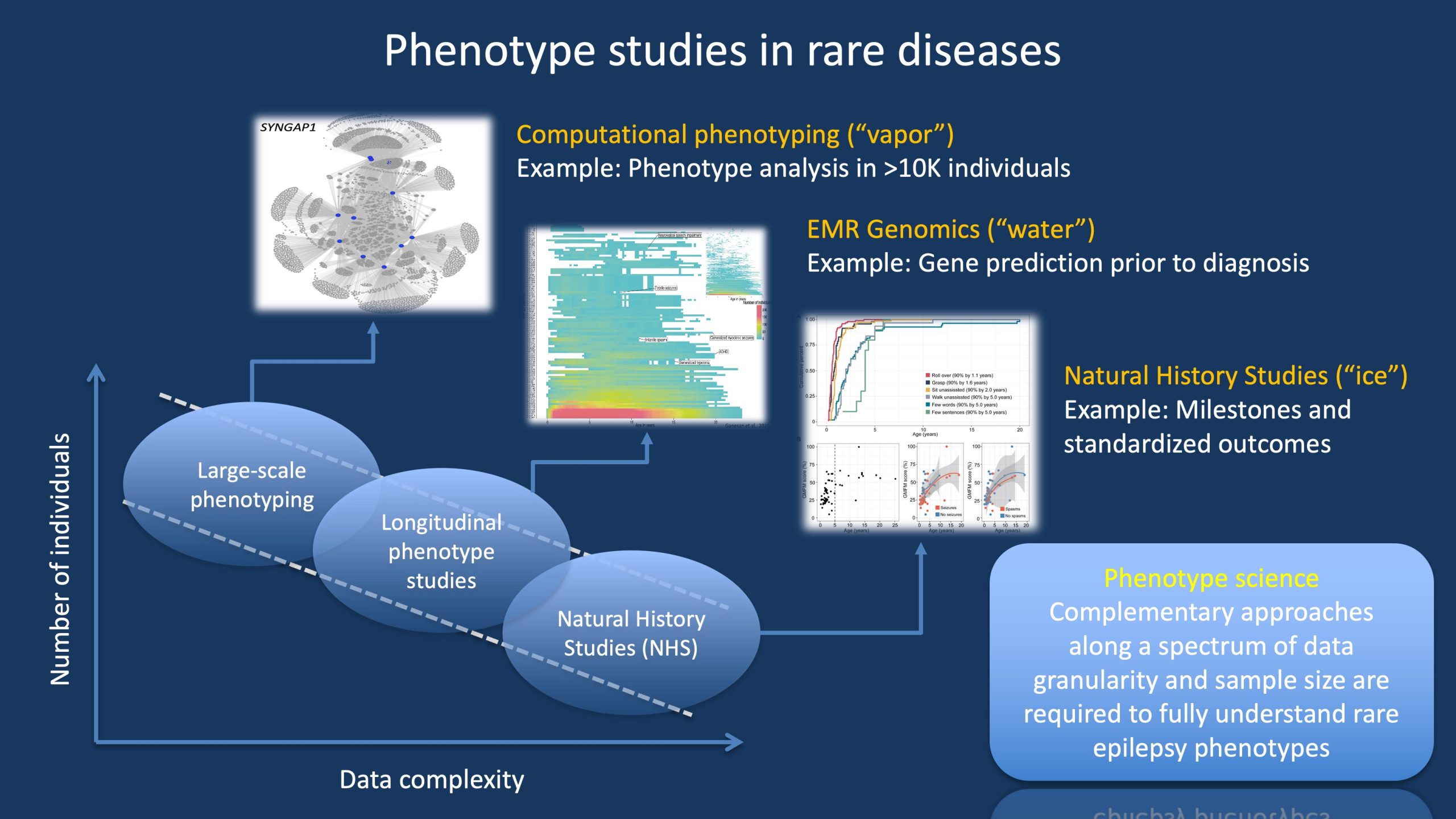
Figure 1. There are different ways to gain understanding of rare disease phenotypes, ranging from approaches with large patient numbers and core phenotypes to smaller studies with highly granular phenotypes. In our blog post, we use the analogy of the different phases of water (ice, water, vapor) to represent the three main approaches to phenotype studies. Importantly, these approaches are not mutually exclusive but complement each other. For example, Natural History Studies and clinical trial readiness studies would be limited if longitudinal phenotype studies were not available to provide a broad overview of the disease trajectory. However, in turn, these longitudinal studies depend on large-scale phenotyping studies to provide an overview of broader disease patterns and subgroups.
Trial readiness. Let me start with the following scenario. We would like to treat a rare genetic epilepsy or neurodevelopmental disorder with a novel compound or a repurposed drug. How would we know that this new therapeutic strategy is effective? The answer to this question is not as simple as it might seem at first glance. Many genetic epilepsies have a broad range of clinical presentations, and the developmental progress or reduction in seizure frequency seen within a given time frame may not simply be due to the new medication that is being evaluated–it might simply be due to the natural history of this condition. We know, for example, that many individuals with STXBP1-related disorders or early-onset SCN2A-related disorders will eventually have reduction or cessation of their seizures, likely unrelated to the underlying medication. However, it is important that this variation does not impair our assessment of the efficacy of a given treatment strategy. In order to conduct trials, we need to be trial-ready. Think of trial readiness as an ice floe.
Ice. The solid phase of water is ice. Ice has a very regular pattern with the molecules rigidly apart from one another connected by the hydrogen bonds that form a crystalline lattice. Ice is stable and can be structured into a defined shape. Think of natural history studies as the rock-solid component of phenotype science. We are thinking about standardized outcome measures and established measurements with respect to development. Clinical trial readiness studies may include objective measurements of seizures, through long-term EEG recordings or quantitative EEGs, and standardized assessment batteries for development, such as the Bayley Scales of Infant and Toddler Development. In addition, the realm of clinical trial readiness includes development of disease-specific scales. The CDKL5 deficiency disorder clinical severity assessment (CCSA) is one example of this. Exploring the difference between Natural History Studies (NHS) and Clinical Trial Readiness Studies deserves a blog post on its own, but one thing is pretty much certain. Whatever is measured needs to be solid and reliable. For example, we would like to know that a novel disease-modifying compound for SCN2A-related disorders reduces seizure burden measured by EEG by 30%, leads to an 10% increase in Bayley-4 scores over two years, and is accompanied by significant improvement on the Observer-Reported Communication Ability (ORCA) and the six domains of the QI Disability measure. Solid as ice.
Water. Standardized outcome measures and clinical trial readiness may be the most tangible endpoints, but what more is there beyond? In fact, this is only a single facet of how we can zero in on rare disease phenotypes. Let me use the following analogy to explain that there is much more to phenotype science. Think of the solid and firm nature of clinical trial readiness as an ice floe. Yes, we are looking at ice, but we zoom out and realize that the ice floe is drifting on a larger body of water–a flowing river. Our ice floe is small compared to the width and depth of the river and is carried by the currents. Phenotypes are more than just the ice-like, solid structure of clinical trial readiness and natural history studies. When we allow phenotypes to transition to a more fluid state, removing them from the crystalline structure of standardized outcomes, we realize that there is much, much more to examine. Standardized outcome measures can only be performed in a relatively small number of individuals, but if we allow our phenotypic molecules to be more dynamic and flexible, we realize that there is an entirely new dimension to evaluate. We are entering what I typically refer to the realm of classical longitudinal EMR genomics. The attribute “classical” is somewhat tongue-in-cheek as this type of phenotype science is relatively new. However, this realm compares to clinical trial readiness studies like ice compares to flowing water. It is less structured, but so much larger, like our river that carries away the ice floes. For example, there is the common misperception that we have little data on rare disorders. This really applies only to the somewhat crystalline or fixed perception of clinical trial readiness. If we look at our medical records, the amount of more fluid information is vast.
The water phase – examples. In our 2020 publication on rare genetic epilepsies, we included more than 3,400 patient years of observational time for rare genetic epilepsies. This data is like water–it does not provide the solidity of firm phenotypes. But it provides an important dimension that clinical trial readiness studies cannot provide–the flow of time. For example, this approach enabled us to find that KCNT1-related disorders are the only genetic epilepsies with a significant risk of spastic tetraplegia by the age of 12 years and that SCN1A-related disorders are most recognizable between the age of 1-3 years. Likewise, these types of studies allowed us to reconstruct seizure histories on a monthly basis for STXBP1-, SCN2A-, and SCN8A-related disorders, emphasizing how the temporal pattern of seizures and prescribed medications deviate over time. EMR-based comparative effectiveness studies show the benefit of the ketogenic diet in STXBP1-related disorders but not SCN8A-related disorders, and analysis of EMR data compiled by medical data aggregators demonstrate that the median age of walking for individuals with STXBP1-related disorders is 3.5 years. These are all examples of the “fluid phase” of phenotype science. This information typically does not replace information in traditional Natural History Studies and clinical trial readiness studies but rather complements it. Or, to stick with our analogy, it represents the large river or underlying current that carries the ice floes.
Vapor. Let me take this analogy one step further. You sit at the side of the river, watching the flowing water and the ice floes that are carried away by it. Suddenly, the entire scenery is cast in shadows and a few minutes later, you can appreciate reflection of the sunlight on the water again. You look up and see clouds in sky that cast long shadows of the landscape in front of you. Clouds consist of condensed vapor, and you realize that there is much more spaciousness and dimensionality to phenotypes when you look at the third phase of water: gas. Phenotypes can also be analyzed when they are presented in a volatile, ephemeral, or momentary state. This is the realm of core phenotypes. Core phenotypes represent the minimal, elemental information that captures the overall essence of an individual’s phenotype. Think of the 6-8 phenotypic attributes that we add to a test requisition, or the clinical report form for large-scale genetic studies. I absolutely agree that these descriptions do not capture the full clinical presentation at all. They lack the solidity of standardized measures or the flux of the disease trajectory, they are elemental, minimal, and the water molecules are not bound together. But why should we think of the gaseous phase as helpful when it comes to rare disease phenotypes?
The gaseous phase – examples. Again, the answer might be surprising. Looking at the minimal core phenotypes allows us to capture patterns that even EMR genomics is too small to identify. For example, we demonstrated that using the core phenotypes of the Epi4K, EuroEPINOMICS, and additional trio exome studies in genetic epilepsies, we were able to reconstruct the phenotypic profile of Dravet Syndrome, even though none of the individuals had been clinically diagnosed. In addition, we were able to identify AP2M1-related disorders as a novel neurodevelopmental disease given the close resemblance of phenotypes and discover that the recurrent SCN2A p.R853Q variant represents a mostly homogeneous disease within a disease based on clinical features. Likewise, even the clinical recognition of Rett Syndrome could be reconstructed through algorithms analyzing these core phenotypes, replicating the clinical insight by Andreas Rett when he first recognized the syndrome that would later be named after him. I typically refer to studies involving these vaporous core phenotypes as Computational Phenotyping studies to delineate them from EMR Genomics and Natural History Studies. The amount of data that is available for these studies even dwarfs the already sizeable data in EMR Genomics. We are currently analyzing core phenotypes coded in Human Phenotype Ontology (HPO) format in more than 10,000 individuals with trio exome and genome data. These phenotypic clouds already provide important insights on the broader patterns within neurodevelopmental disorders and genetic epilepsies.
What you need to know. There are different ways to think about phenotypes, which are mutually complementary. In brief, we need ice, liquid water, and vapor. While we frequently think about phenotypes assessment mainly as Natural History Studies in the era of precision medicine, EMR Genomic Studies (water) and Computational Phenotyping studies (vapor) are necessary complementary approaches to direct the more detailed and in-depth assessments required for trial readiness. Only by jointly looking at the three phases of the phenotypic substance will we be able to get a complete overview of the clinical features in rare genetic epilepsies. The integration of these three phenotypic states will be important for translating our understanding of disease phenotypes into treatments.


