Precision medicine. This post continues the discussion on how we can make sense of clinical data in the absence of outcomes in the context of precision medicine – a concept that drives much of what we do on a research basis. The fundamental idea is that clinical care in pediatric epilepsies can be personalized and tailored to underlying etiologies. With continual progress in gene curation and variant interpretation alongside clinical knowledge, we typically expect that treatment suggestions are immediately implemented after the discovery of the causative genetic etiology. For example, a child with early onset epileptic encephalopathy is found to have a gain-of-function variant in SCN8A and is almost immediately started on a sodium channel blocker such as Trileptal. However, to what extent is this the case? In the context of precision medicine, how precise are we exactly?
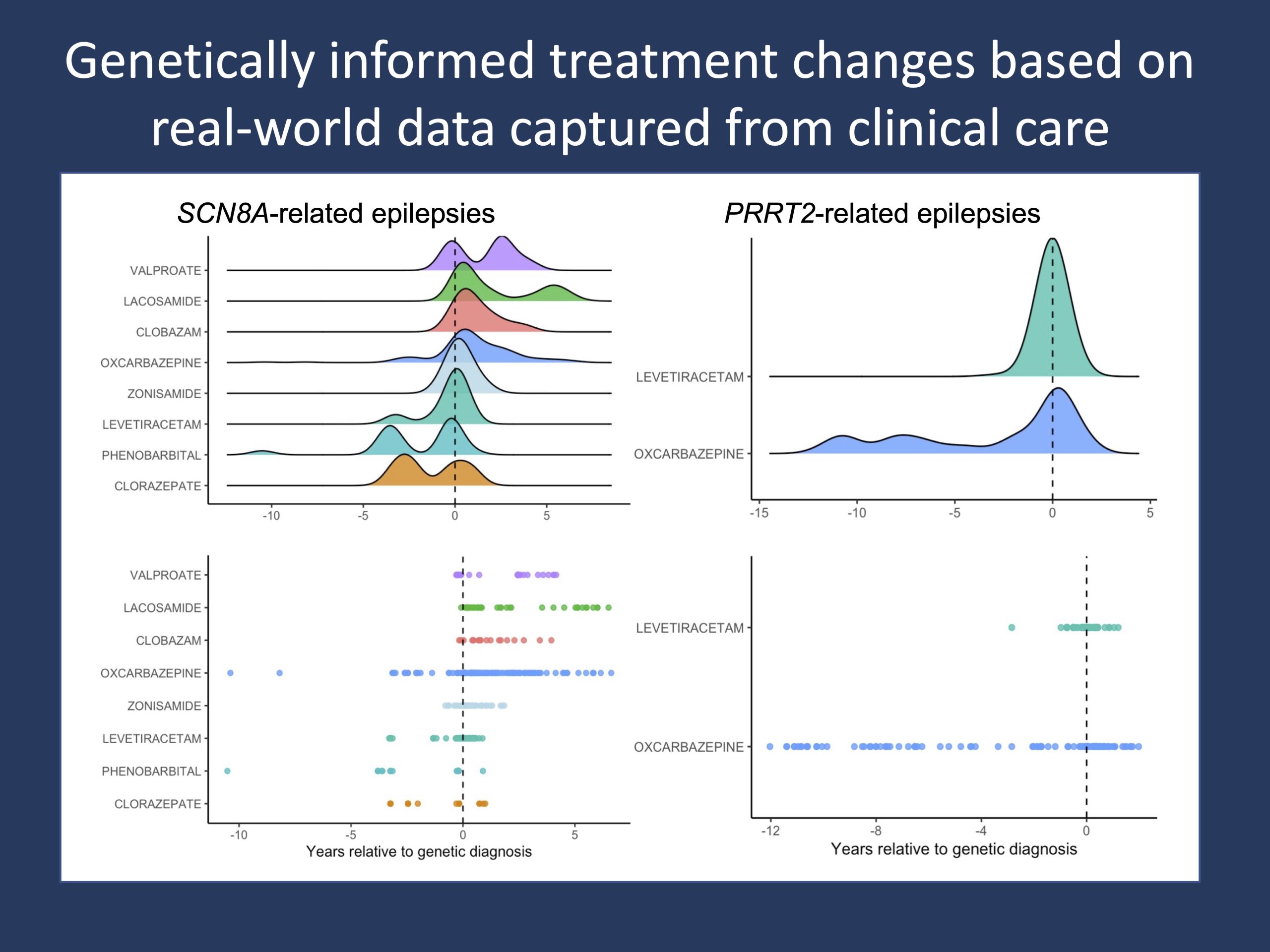
Figure 1. Changes in anti-seizure medication (ASM) prescriptions in temporal relation to a genetic diagnosis in SCN8A-related disorders and PRRT2-related disorders. ASM data prescribed as a part of routine clinical care are systematically captured at scale in the Electronic Medical Records (EMR) and can be assessed in the context of recommended treatment strategies tailored to genetic etiologies, such the initiation or discontinuation of certain ASMs in specific epilepsies (Symonds et al., 2019), reflecting the degree to which real-world decision-making and clinical practice align with consensus and theoretical guidelines in epilepsy management.
Treatment guidelines. In genetic epilepsies, it has become almost universal knowledge that sodium channel blockers such as oxcarbazepine and phenytoin are contraindicated in Dravet Syndrome in individuals with loss-of-function SCN1A. On the flip side, these same drugs are recommended in the setting of gain-of-function (GoF) variants in epilepsy-related sodium channelopathies such as in individuals with the recurrent p.Arg1872Trp/Gln/Leu SCN8A variants. Thus, when thinking about evidence-based medicine, there are well-established frameworks and a generally agreed upon consensus understanding for informed treatment choices in the epilepsies. However, to what degree do these guidelines translate into real-world practice?
Real-world data. The brief answer is that real-world clinical care is highly variable and that genetic diagnoses do not always translate into evidence-based changes in treatments. When assessing anti-seizure medication (ASM) changes following a genetic diagnosis in a cohort of >2,500 individuals with genetic epilepsies, we find that treatment strategies are more heterogeneous than we might think. While we observe expected ASM changes such as the initiation of oxcarbazepine and lacosamide in SCN8A-related disorders and oxcarbazepine in PRRT2-related disorders (Figure 1), we also capture more commonly and broadly used ASMs such as levetiracetam emerge as significant choices following a genetic diagnosis. This heterogeneity can be seen to a much greater degree in other epilepsies. A particular example that we have recently looked at through a project funded by the NORSE Foundation is the rapid administration of anti-seizure medications in refractory status epilepticus and FIRES, where a range of medication sequences and combinations are given.
Granularity. The philosophy of using Real-World Data (RWD) is that we can capture and better understand these nuances from longitudinal data generated through routine clinical care. In fact, the granularity of this data is documented in the Electronic Medical Records (EMR) to the level of a minute timescale. In the setting of refractory status epilepticus, we can assess anesthetic infusions administered with escalation in dosages and medication status by the minute. However, even with this level of granularity in our data, why is this important if we do not have defined outcomes to compare to?
A prior hypothesis. The systematic assessment of clinical data allows us to measure and quantify clinical decision making. Independent of outcomes, it helps us understand what we are doing on a clinical day-to-day basis. As the administration of ASMs are captured in a standardized record database and agnostic to provider interpretation, it permits an unbiased view of the dynamic treatment landscape at play within a large healthcare network. In the absence of outcomes, we can still make clinically meaningful conclusions such as the impact of a genetic or molecular diagnosis on patient care. That is, even without seizure data relative to treatment, we can characterize how a diagnosis affects the clinical management of patients and quantify the degree to which clinical management aligns with established consensus guidelines. So, how does this connect back to precision medicine?
Learning health system. The basis of a learning health system starts with the aggregation and analysis of data captured from routine clinical care. The knowledge that is generated then informs the development of appropriate interventions and is eventually translated back into patient care. While there are long-established and clear guidelines in the field with tailored treatment recommendations for conditions with known etiologies, it is more often the case that clinical care is carried out with more heterogeneity. In FIRES, it is not uncommon that the second line anticonvulsant is not given within the hour following administration of the benzodiazepine as per consensus guideline. This analysis is not limited to treatment data, and other modalities of care can be assessed such as the diagnostic gap – the time between first seizure onset and genetic testing sent. The first step, however, is to characterize the real-world landscape.
Conclusion. The degree to which theoretical guidelines are translated into clinical practice varies. The systematic and large-scale analysis of real-world data is essential in providing valuable insight into the extent of which precision medicine efforts are being implemented – even without clinical endpoints. However, more importantly, it gives insight into what we are doing, and it allows us to critically assess the domains in which clinical care can be more precise. Given that treatment data will be available at much larger scale than outcome data, information on changes in clinical practice after a genetic diagnosis may in some cases be the best information out there to guide future treatment, even in the absence of outcome data.


