Zebra finches. Exactly one year ago, I wrote my last blog post on the genetics of stuttering and thought that it would be time for an update. Here, I would like to explore why stuttering is a truly neglected neurogenetic disorder and why we have made so little progress. In addition, I would like to give a brief update on where we are right now, looking at stuttering from the perspective of the wider pediatric neurogenetics field. In addition, we will unleash the power of EMR genomics to query the medical records of more than 52,000 individuals to find associated genes, and we will discuss a monogenic cause of familial childhood-onset fluency disorders that we did not expect to find. Here is a summary of the last 12 months in stuttering genetics.
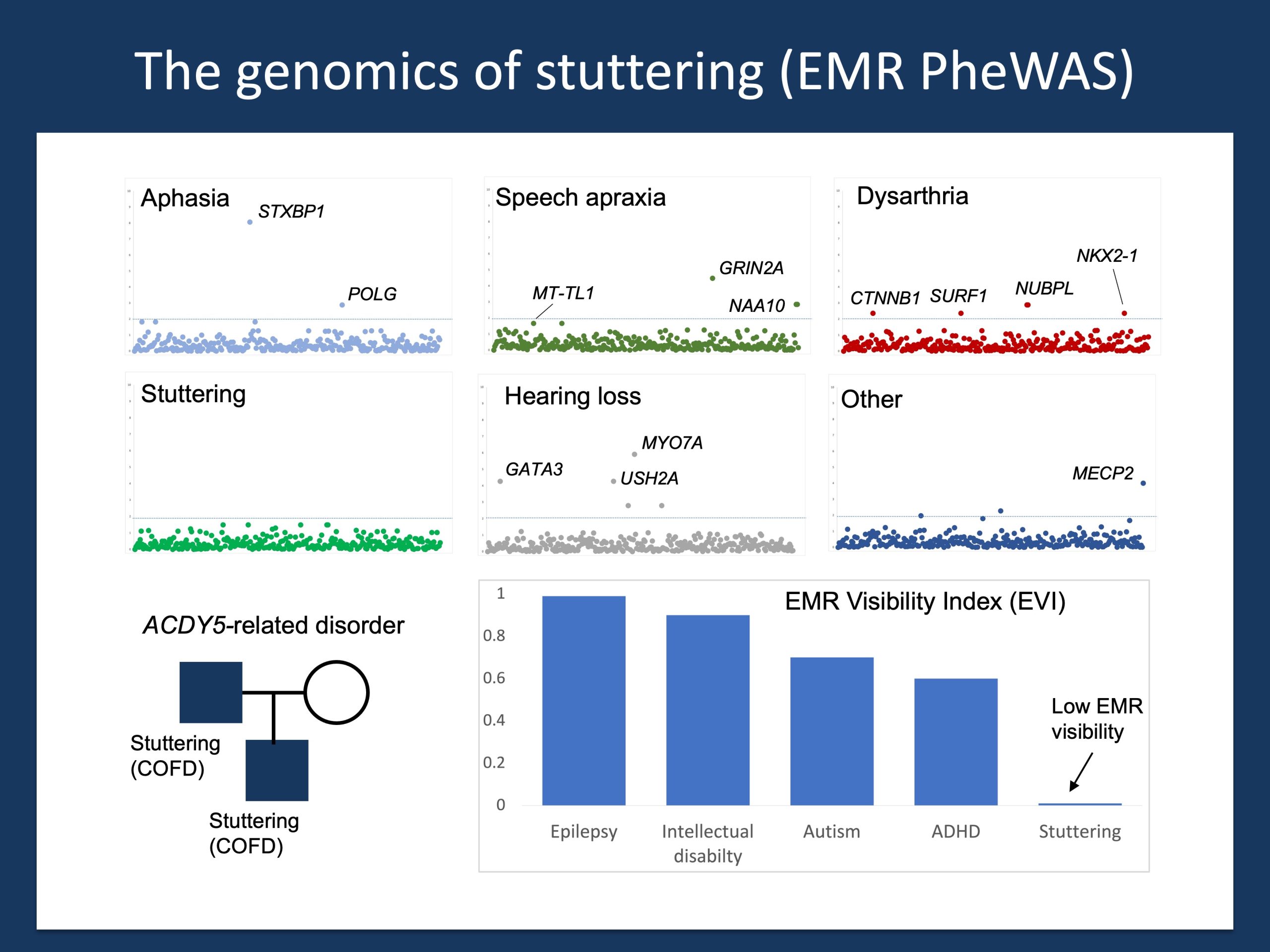
Figure 1. EMR genomics in childhood-onset fluency disorder (COFD) aka stuttering. Understanding the genetics of stuttering is not trivial. We searched the electronic medical records of more than 52,000 individuals with speech disorders for associated genetic diagnosis. While we find known associations such as aphasia (non-verbal) with STXBP1 and speech apraxia with GRIN2A, none of the almost 300 genetic etiologies we identified in the patient notes were found in more than one family with stuttering. The only known genetic cause was found in a family with a variant in ADCY5, a genetic cause for a wide range of movement disorders. The reason why stuttering is difficult to identify in existing biobanks and genomic resources is shown at the bottom right. Stuttering has a very low “EMR visibility”. Most individuals with stuttering do not carry the diagnosis in the medical records, which leaves us blind when trying to mine the massive resources such as the UK Biobank or FinnGen for genetic links to stuttering. This is not the end of world, stuttering is simply the first true postgenomic disease that requires a novel approach.
My own story. Stuttering or “childhood-onset fluency disorder” (COFD) is my very own neurodevelopmental disorder. Therefore, as many other people with speech disorders, I have my own story to tell. A few weeks ago, I participated in podcast by the Nolan Foundation (link) where I told my very own story, ranging from the time my elementary school teacher asked my class not to tease me about my stutter to the absurd (and funny) situation of working in a high-powered neuroscience lab in Heidelberg that discovered most of the GRINs and GRIAs and being unable to say “glutamate” (even today, I say “ga-lutamate” to overcome dysfluencies).
Superpowers. Don’t get me wrong, I am not telling a sad story in this Nolan Foundation podcast. At some point, I am even commenting on the stuttering superpower. This is a skill that many stutterers and I have developed as a consequence of our dysfluencies, namely the rapid ability to pick up the mood in a room faster than anybody else. Of course, it is not really a superpower, but a phenomenon that is increasingly referred to as the stuttering benefit. Speech disorders are disabilities, and many individuals with dysfluencies in a fluent world develop compensatory skills. When I was younger, I always felt that I had “antennas” that nobody else had. But I don’t want to digress, let me get back to my genetics update for 2023.
Our team. Understanding the genetics of speech disorders is not something that we can do in isolation. I am very happy and honored to be part of a team that we have built over the last two years. Jan Magielski is our Penn student working on this project, Sarah Ruggiero is our genetic counselor who holds everything together, Joe Donaher runs our local stuttering program and is our resource behind everything we have learned about stuttering, and Alex Gonzalez navigates our electronic medical records (EMR) like nobody else. It has taken some time to build this focus of our group, and this is a big shout-out and thank you to our team. For anyone reading this who is interesting to join our stuttering genetics team, please let us know – there is hardly anything more exciting in speech genetics at this point.
Stuttering genetics. Let me say upfront that our biggest achievement is that we have not given up. I have had several false starts in my life trying to get involved in stuttering genetics and not giving up is a VERY big thing given the shame and guilt that is typically associated with dysfluencies. While we have not been as fast as we wanted to, we have made progress. We have successfully expanded our epilepsy and neurogenetics research protocol to include stuttering (after all, it is a neurogenetic disorder), and we have slowly, but consistently recruited participants into our study. Getting genome sequencing will be next, taking one step after the next.
Pediatric neurogenetics. We have also taken our very own approach to dissecting the genetics of stuttering, taking advantage of the tools that our team has developed. We can dig deep into electronic medical records of large patient cohorts, and we have genetic information on a large group of children with neurodevelopmental disorders. Trying to understand the genetics of stuttering through the lense of pediatric neurogenetics is one worthwhile approach that we are pursuing. For example, in 2022, the team by Angela Morgan and collaborators identified a unique stuttering phenotype in children with KANSL1-related disorders, a condition previously known as Koolen-de Vries Syndrome. Findings like this help propel the field forward in the absence of large, available data sets. But why are these datasets lacking at all, given the large amount of information available through large biobanks such as the UK Biobank or FinnGen?
EMR Invisibility. The answer is relatively easy and disappointing. Stuttering is EMR invisible. In a pilot study, we compared the frequency of clinical diagnosis mentioned in the full text clinical notes and the presence of and ICD10 diagnosis code (Figure 1, lower right). Many children with stuttering mentioned in their patient notes do not have stuttering listed as a diagnosis. As many biobanks use the ICD10 billing codes rather than translated fulltext notes, this leaves stuttering EMR-invisible. There is absolutely NOTHING on stuttering in the large biobanks that are publicly accessible. What can we do to overcome this issue? We can look at large clinical datasets using a different approach.
EMR PheWAS. Let me introduce you to the concept of an EMR-based phenome-wide association study (EMR PheWAS). Imagine the following: we systematically comb through all medical records, identifying individuals with diagnosis codes for various speech disorders and at the same time screening the medical records for the mentioning of a wide range of possible genetic diagnoses. This is what we did to understand the lay of the land. Our preliminary results, where we screened the medical records of more than 52,000 children with speech and communication disorders, is shown in the upper panels of Figure 1. This analysis naturally happens in broad strokes, and there are many, many things that likely escape our attention. However, one result became clear very quickly. None of the 290+ genetic diagnoses for which we were able to screen the EMR were found in two or more individuals with stuttering/COFD. There does not seem to be a common pattern of neurogenetic disorders that is as prominent as the association of GRIN2A and speech apraxia or STXBP1 and aphasia (a proxy term for “not communicating with spoken language” in the EMR). When compared to five other speech or communication diagnosis codes, this absence is striking, adding to the ongoing mystery of stuttering.
ADCY5. One finding is an exception. We identified a single family with an autosomal-dominant inherited variant in ADCY5 with two affected individuals with a stuttering phenotype. This finding was surprising. ACDY5 is not a typical neurodevelopmental disorder, but a genetic etiology linked to movement disorders, the so-called ADCY5-related dyskinesias. While this was only identified in a single family, genes for movement disorders are likely underdiagnosed in stuttering families. Making such a diagnosis is important, as at least some of the genetic movement disorders have established treatment pathways. In addition, more generally, this preliminary finding supports the notion that at least some aspects of stuttering are movement disorders. Thereby, stuttering is closer to conditions such as tic disorders and Tourette Syndrome than speech apraxia. This puts a renewed focus on the basal ganglia (movement disorders) as opposed to cortical structures (speech apraxia). We will see in the future whether other genes for movement disorders can be found in additional families with dysfluencies.
What you need to know. There is steady progress in stuttering genetics, and we are systematically chipping away at the barriers that prevent us from having a better understanding of the underlying genetic basis of stuttering. Being able to slowly bring stuttering genetics to eye level with other neurodevelopmental disorders cannot be overstated. Our goal is not to make genetic diagnoses to label individuals and thereby “other” people who stutter (after all, I am a person who stutters myself). In contrast, we hope for a fundamental understanding on why dysfluencies happen as a feature of human diversity. There are many myths and private origin stories on why people who stutter have acquired their dysfluencies, be it the hit on the head, the mean older sibling, or a presumed underlying anxious personality. Many of these factors have been studied and disproven. I hope that understanding the genetic basis of stuttering can help us resolve many of the misunderstandings about the causes of stuttering that often weigh heavily on people who stutter and their families. We stutter because of our very own genetic make-up, and this is simply a part of what makes people different. And this information may hopefully help us reinforce the fact that there is no diversity without dysfluency.


