Data sharing. Over time, genomic scientists have learned how to share. Large international cohorts and efforts of data deposition have led to large databases that can be used to answer big questions. However, silos of genomic data, such as massive sequencing studies performed on specialized cohorts, lay unconnected across research groups, academic institutions, and collaborators. Recently, we have been involved in several projects to de-silo rather than simply share genomic data, and we realized that there may be some aspects that apply to the genomics worlds more broadly. For example, what makes de-siloing different from data sharing? The goal of this post is to redefine these concepts and explain why we should be less concerned about data sharing and more concerned about data integration.
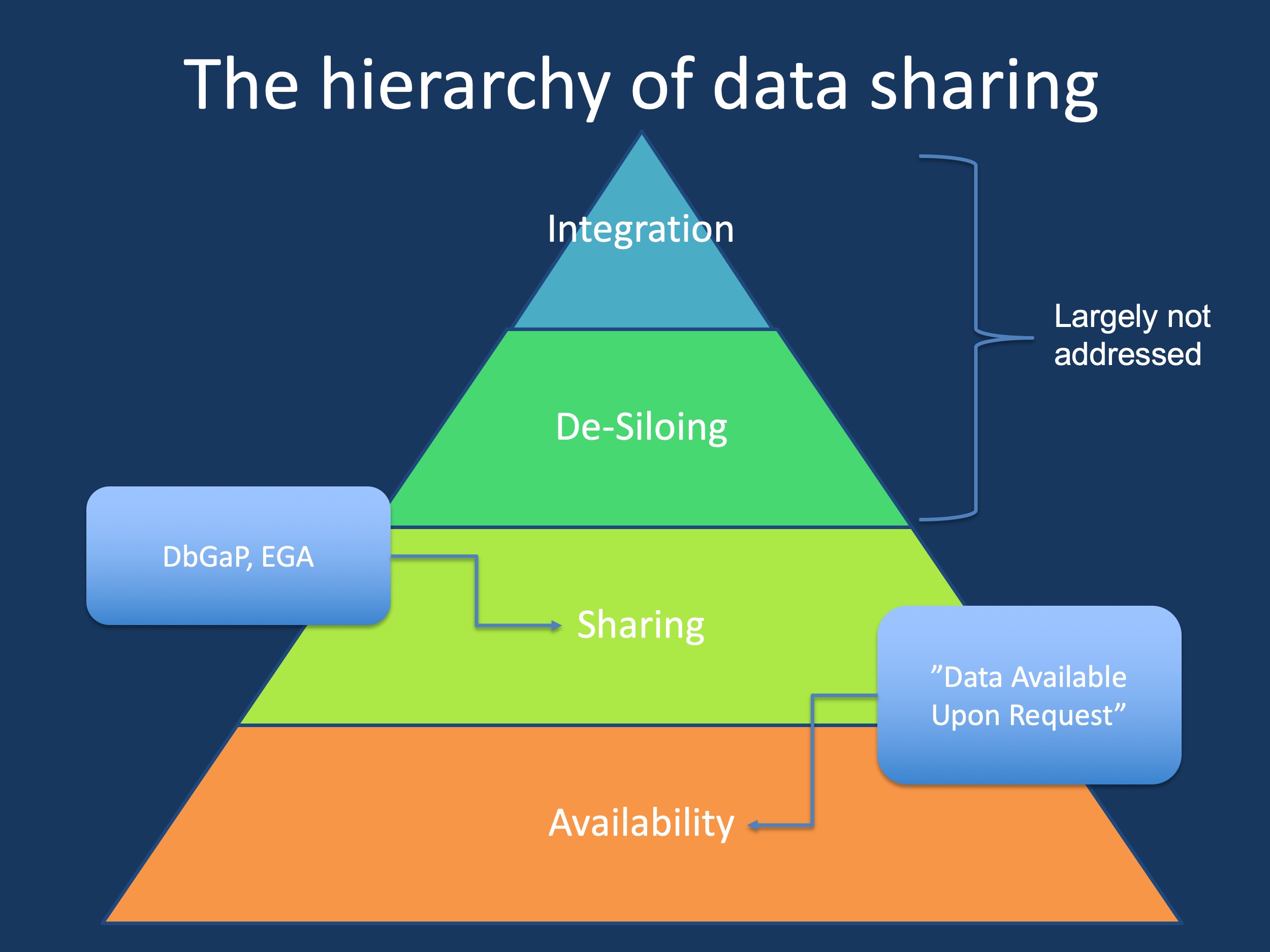
Figure 1. The data sharing pyramid. Intentions to provide access to research typically starts with a commitment to data availability. The next higher level is data sharing, typically in a minimal format into databases. De-siloing and data integration, the topics of this post, are still different from this as they require an active effort from the recipient to retrieve and harmonize data.
Does data sharing work? We know that this is a provocative question as data sharing is currently a hot-button issue in the genomics world. Everybody is in favor of data sharing and nobody is really against it – if you ask a crowd of genomic scientists about their attitude towards data sharing, you will most likely get an enthusiastic YES. But what really happens after this? Let me give you some examples why we are trying to disentangle the complexity of data sharing and why we are trying to use somewhat different words and concepts for this.
The format. Here is one example of our 2020 study on phenotypic signatures in genetic epilepsies. We combined data from various consortia for this study. Obtaining genomic data was easy – deposition is standardized through resources such as dbgap and the European Genotype Archive (EGA). However, retrieving the already databased clinical data took us almost six months – we needed new data sharing agreements and significant time and effort on our end. For some datasets, we never received the phenotypic information. All this data was supposedly readily available for secondary research.
Available upon request. Most of us have dropped this line into manuscripts in the past: “Data available upon request”. However, while this statement often satisfies reviewers and editors, it is largely ineffective. A recent study reports that more than 90% of studies including such a data availability statement do not share data in the end. There might be a wide range of reason for this. For example, data transfer between institutions becomes increasingly complex and requires agreements. In addition, there is often a back and forth about data formats and both parties may have different standards or technological solutions. For example, is every lab immediately possible to retrieve 1000+ genome BAM files? However, in the end voluntary data sharing rarely happens. In fact, in our epilepsy study, the community truly overperformed – for most studies, we ended up retrieving the data that we needed.
Persistence required. So, what goes wrong with data sharing? The answer is relatively straightforward. There are simply no meaningful incentives to share data. There are no metrics and no rewards. Therefore, in a competitive research environment, the lack of data sharing is to be expected. Various solutions have been tried in the past. Genomic data sharing is mandatory for NIH projects and some projects such as the Gabriella Miller Kids First Projects specifically generate data for broad secondary use. But there are other strategies – one strategy is de-siloing.
Sharing versus de-siloing. Why are we trying to emphasize the difference between these concepts? In brief, from the perspective of the data recipient, data sharing is passive whereas de-siloing is active. De-siloing is an activity by the data recipient to maximize the use of the available information, actively reaching deep into data silos to extract as much meaningful information as possible and thereby shaping the process. Data sharing, in contrast, is passive – you get what you get. It goes without saying that de-siloing requires time and effort, it is an activity not unlike data analysis itself. ETL (“extract, transform, load”) is one concept related to this, but does not capture the entire complexity of genomic data. To make a long story short, we have gained quite some experience in data de-siloing.
De-siloing versus data integration. As with sharing versus de-siloing, we are drawing an artificial line, especially as the data integration process in the genomics sphere requires additional considerations. Data harmonization is critical for genomic data that was generated across different projects and genome builds and may have been saved in various formats (CRAM, BAM, FASTQ). Phenotypic data is more complicated – one major activity of our lab is phenotypic data harmonization through biomedical ontologies, such as the Human Phenotype Ontology (HPO). Basically, we are trying to use common dictionaries to make sure that the fact that “absence seizures” are a type of “generalized seizures” is preserved. In our prior work, we have seen that the amount of clinical data increased 5-10x through such processes. And for specific datasets, information only really becomes useful once it is harmonized and integrated.
So, what is the future of data sharing?We would argue that data sharing only really works if the receiving party actively uses the data and shapes the process. Large data repositories will always exist, but if we would like to meaningfully use previously generated data in its fully extent, data sharing cannot remain a static process of data deposition, but will need to require ongoing, active curation and refinement. Therefore, we might soon live in a future where the genomics community values and perfects the art of de-siloing rather than just data sharing.
This blog post was co-written by Sarah Ruggiero and Ingo Helbig.


