Classification. Our classification of the epilepsies periodically undergoes revision to align the way we think about the epilepsies with scientific progress in the field. While it is intuitive that relatively novel frameworks such as the 2017 International League Against Epilepsy (ILAE) Operational Classification of Seizure Types capture the current spirit of the field more accurately than prior classifications, one relatively simple question is not easily answered: how much more accurate? How we get to such an answer requires us to take a step back and think about how the value of clinical information can be measured and compared. In our recent publication, we describe the revision of the Human Phenotype Ontology (HPO) according to the most recent ILAE classifications and other respected definitions in current use. This gives the answer to the prior question: 40% (which is a lot).
HPO. The Human Phenotype Ontology is a controlled dictionary to map 16,000 clinical features within a single framework. In our recent study (Lewis-Smith et al., 2021), we revised the seizure subontology to align it with the most recent ILAE classifications. We compared how comprehensively the same seizure data could be captured through this revised framework rather than a prior HPO version based on previous clinical concepts. We find that the overall information content increased by 40%.Data harmonization. The idea behind the HPO is best described using an example. If you would like to combine clinical data collected alongside Epi4K and EuroEPINOMICS, two of the early major epilepsy genetic studies, how would you accomplish this? While exome data can easily be combined (as we did within the E2 initiative), combining the phenotypic data collected in various data formats is difficult. This data needs to be harmonized to make it comparable, making sure that field x in one dataset correctly corresponds to field y in the other dataset. Alternatively, we could translate both datasets to a common standard, ideally a standard that is in widespread use in other areas in genomics, and freely available. The HPO is emerging as such a standard and is used in a variety of research projects and clinical services, including the DDD study, the UK’s 100,000 Genomes Project, the NIH Undiagnosed Disease Network, the GA4GH, and hundreds of global initiatives. The HPO also contains a rich vocabulary for epilepsy-related phenotypes, which we have developed in prior large research networks, and which we continue to expand. Whether or not a genetic variant of uncertain significance is reported out is increasingly dependent on the HPO, as it is used by many diagnostic laboratories.
What the HPO is good for. Once data has been mapped and harmonized to the HPO, information can be analyzed jointly – and it grows relatively quickly. Most of our lab’s recent projects have taken advantage of the HPO and some of the results emerging from this analysis are quite interesting. For example, the relatively limited clinical data in Epi4K and EuroEPINOMICS is sufficient to reconstruct the clinical features of Dravet Syndrome, even though none of the participants had been diagnosed clinically, it allows us to find new genes such as AP2M1, and previously hidden clinical subgroups in conditions such as SCN2A-related epilepsies. In brief, using the HPO allows us to finally overcome some aspects of the phenotypic bottleneck that have held us back for so long. Also, an HPO-based framework is a useful tool to map the heterogeneous and sparse clinical data from the Electronic Medical Records (EMR), one of the next big frontiers in epilepsy research.
Improving the dictionary. When applying controlled dictionaries such as the HPO, one major issue arises. The quality of the data mapping is necessarily limited if the dictionary is incomplete or imprecise. For example, if a dictionary does not allow us to distinguish between focal seizures and generalized seizures, this distinction will be lost. We measure the amount of information captured through a controlled dictionary such as the HPO using the concept of “information content”, which formerly measures the distinguishing value of each term within the dictionary. Accordingly, two competing dictionaries can be compared by measuring the total amount of information captured. The definition of information content, -log2 of the frequency of a phenotypic term, allows for precise definition of clinical concepts that have frequently been used, but which have always been somewhat fuzzy, such as “phenotyping depth”.
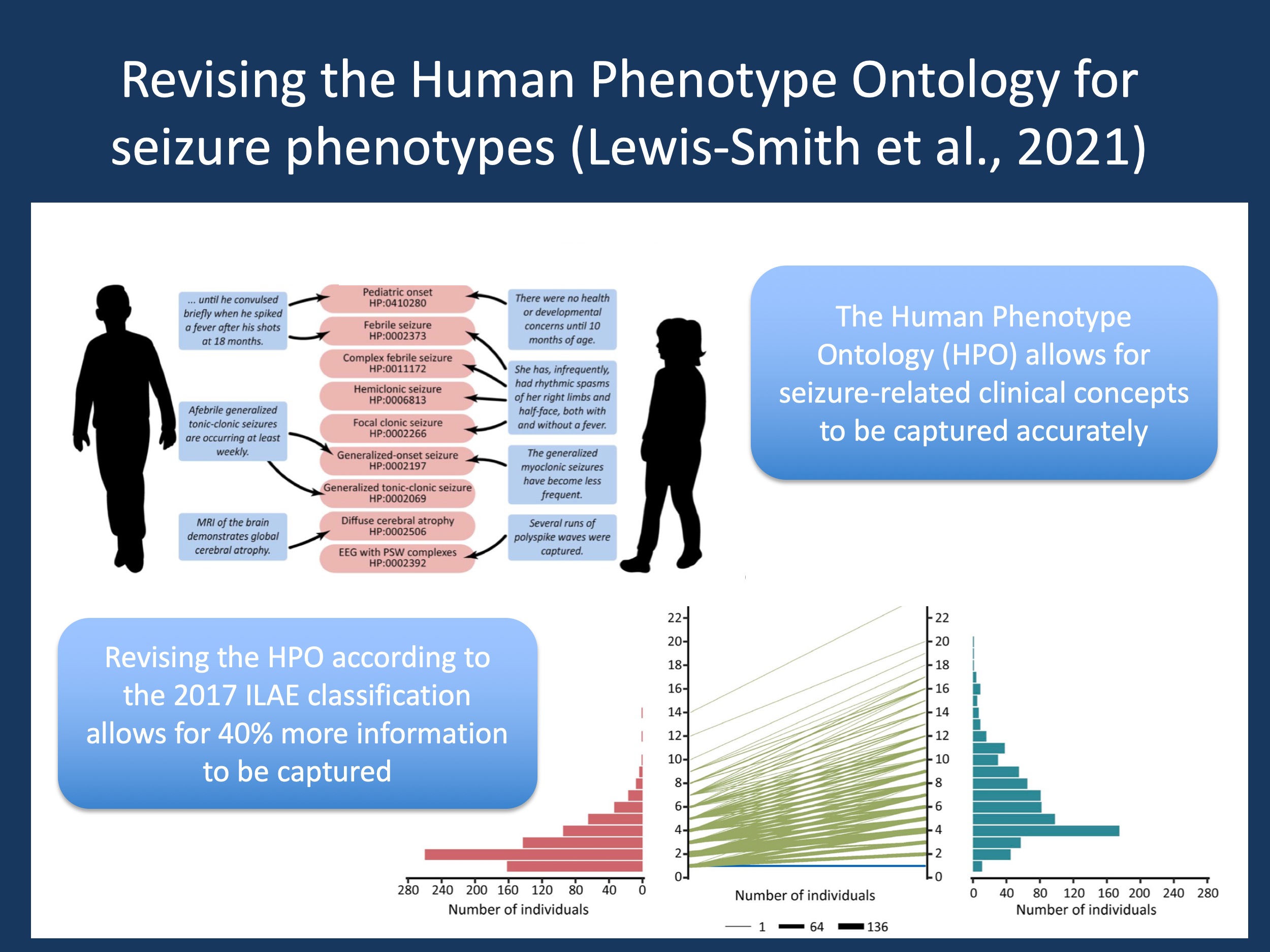
Revising the HPO. In our recent paper in Epilepsia, we describe our process of revising the HPO and our initial experiments to assess how good this new mapping is. In our project that emerged from the Epilepsiome Task Force of the ILAE Genetics Commission, we created a new HPO seizure subontology based on the 2017 ILAE Operational Classification of Seizure Types, and integrated concepts of status epilepticus, febrile, reflex, and neonatal seizures at different levels of detail. Our final subontology, that has been available on hpo.jax.org since December 2020, increased the number of seizure terms in the HPO five-fold, now allowing for a much more precise description of seizure terms. This new subontology is part of the official HPO release and therefore already in use.
Showing improvement. To find out how much better this new ontology performed, we compared the HPO seizure subontology before and after our revision on 791 individuals from three independent cohorts, including individuals we contributed to the Epi25 project. The increase in information content captured by this new ontology was astonishing: the information captured in the cohort increased by almost 40%, as did the number of seizure terms that could be annotated. This comparison now allows us to estimate how well improvements of the HPO might be in related fields, such as autism or cerebral palsy. In brief, the expected improvement is not subtle, its change is significant. While we had previously used the HPO simply as a tool, our study by Lewis-Smith and collaborators now examines how important it is to make such a dictionary more comprehensive, internally consistent, and precise.
What you need to know. In our study by Lewis-Smith et al, we aligned the HPO with the most recent ILAE classifications and other emerging classifications in the field, providing an improved, state-of-the-art dictionary to map clinical features of the epilepsies to a common framework. The HPO is a major language used in genomics and emerging Big Data projects, now allowing us to express important aspects of the epilepsies more clearly, which makes it possible to capture many of the subtle differences between epilepsies that may be relevant to diagnosing and treating our patients.