Natural History. Over the last few years, there has been a renewed interest in outcomes and natural history studies in genetic epilepsies. If one of the main goals of epilepsy genetics is to improve the lives of individuals with epilepsy by identifying and targeting underlying genetic etiologies, it is critically important to have a clear idea of how we define and measure the symptoms and outcomes that characterize each disorder over a lifetime. However, our detection of underlying genomic alterations far outpaces what we know about clinical features in most conditions – outcomes such as seizure remission or presence of intellectual disability are not easily accessible for large groups of individuals with rare diseases. In this blog post, I try to address the phenotypic bottleneck from a slightly different angle, focusing on how we think about phenotypes in the first place.
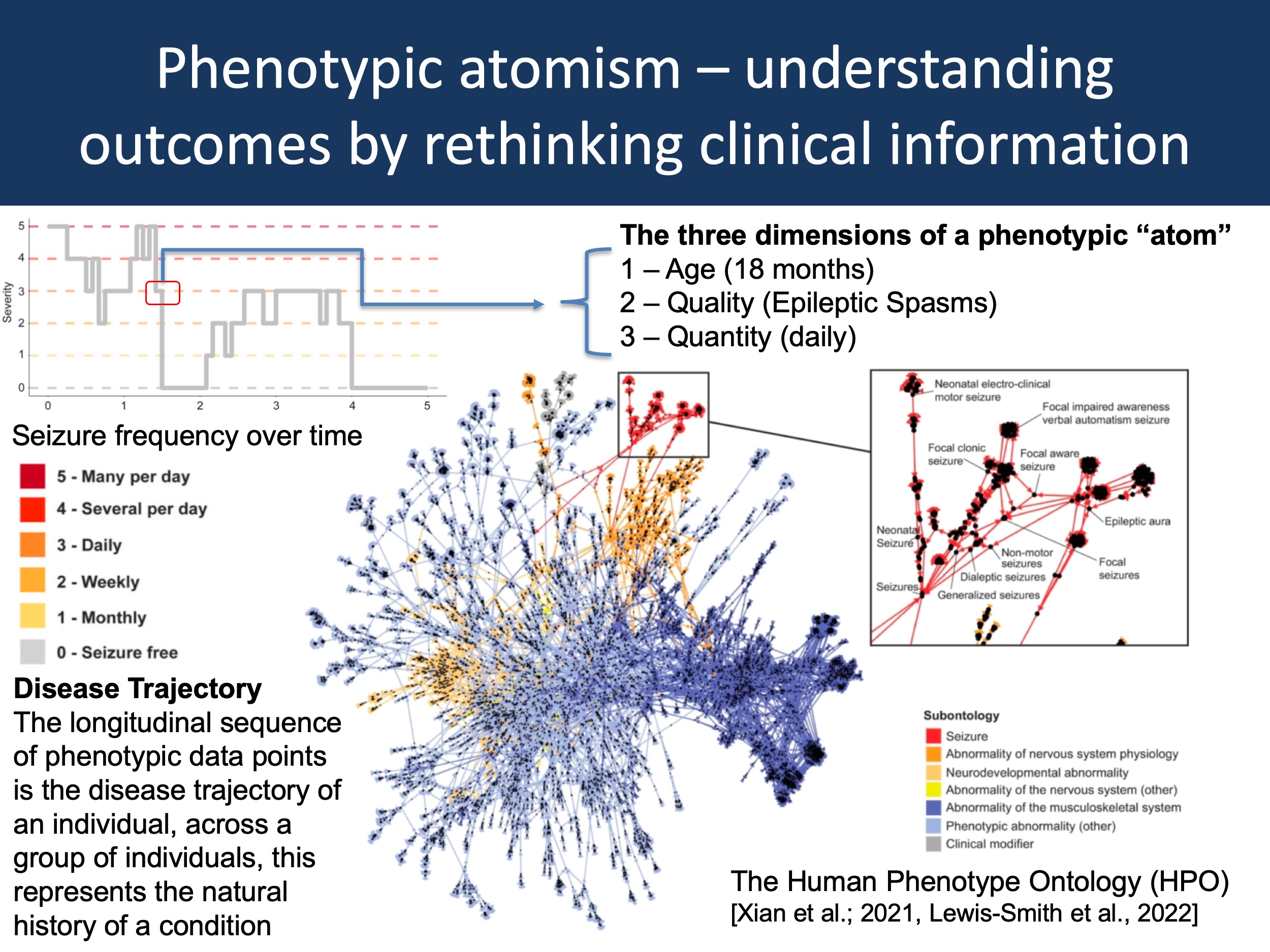
Figure 1. One way to think about a disease trajectory is the description of a sequence of data points with three properties: age, quality, and quantity. In the figure above, I have taken the example trace of seizure frequencies in one individual with epilepsy from Xian et al., 2021 (upper left) to demonstrate this concept: the “atom” is Epileptic Spasms at 18 months with daily seizures. If we accumulate all of these phenotypic “atoms”, we obtain a complete picture of the longitudinal disease history. If descriptions of quality and quantity use standardized format such as the Human Phenotype Ontology (HPO, lower right), an individual’s phenotype becomes comparable with other existing data sets, enabling the construction of disease-specific natural histories.
Atoms. Let me start out with the provocative thought that we typically do not think about phenotypes in the right way. Our approach to assessing the clinical presentation of the genetic epilepsies is often biased and driven by a specific purpose. For case series, we typically like to outline differentiating features. For clinical trials, we focus on measurable endpoints. For medication responses, we distinguish between response and non-response based on clearly defined criteria, as is also the case for treatment-resistant epilepsy. These concepts feel very natural to us as we use them all the time, both in clinical practice and research. However, I would like to argue that something is missing from this approach, namely a deeper understanding of what disease phenotypes really are. Let me take a few steps back to explain.
Next generation sequencing. When SNP arrays, GWAS and NGS technologies first entered the scene, a typical scientific presentation would focus predominantly on the technical aspects, describing and celebrating these novel technologies and their potential. The step change in throughput and standardization of these technologies resulting from these methodological advances yielded novel insights into previously inaccessible conditions. Much of the activity in the field was centered on technology development and using analytical methods to delineate the underlying concepts. For example, the concept of a genetic architecture or missing heritability represent conceptual developments that go far beyond seeing these new technologies only as tools that need to directly demonstrate their utility. We were very patient with these technologies, even if they did not yield immediate benefits. The reason I am giving this example is that I am somewhat lacking the same patience when it comes to understanding the science of phenotypes. I am not referring to specific insight into clinical features of a given disease, but the underlying principles of phenotype analysis more broadly.
Phenotypes beyond outcomes. I sometimes explain to collaborators that our understanding of phenotypes is in the BAC (bacterial artificial chromosomes) era. Before there were SNP arrays and exome sequencing, BACs were a common method to map genomes. For example, the Human Genome Project relied on BACs to sequence the first human genomes. Nowadays, we would use long-range sequencing and de novo assemblies. What I mean by this comparison is that our understanding on how phenotypes are conceptually structured is in its infancy. Our methods to assess phenotypes are still relatively blunt and the underlying concepts not well developed. And in the same way that an early-day NGS expert would have asked you to patiently listen to a description of the methodology and the total amounts of base pairs sequenced, let me try to build up a conceptual framework of phenotypes in the same way. Yes, I will arrive at outcomes and natural histories, but let me start with phenotypic “atoms”.
Machine learning. Clinical data is abundantly available, and our problem is not the existence of clinical information but our ability to access and use it. In the current age of large computational resources and artificial intelligence, the thought that algorithms can comb through the millions of patient records and use deep learning techniques to help us understand what medications work best for individuals with Dravet Syndrome or SCN2A-related disorders is tempting. We are often made to believe that data just needs to be sufficiently large, and then the answers will emerge from the noise. I would like to refer to this viewpoint as machine learning idealism, though yes, this might be absolutely possible in the future. According to this view, data can remain unstructured and underlying larger frameworks on how clinical information can be conceptualized are unnecessary and potentially even harmful. Here, I would like to suggest an opposing view, contrasting machine learning idealism with phenotypic atomism. Basically, I am trying to explore the implicit concepts that our group has developed on how we think about longitudinal clinical data.
HPO. Over the last few years, we have used the Human Phenotype Ontology (HPO) in much of our work. However, the idea of phenotypic atomism is not about HPO. In fact, there is often a misunderstanding when it comes to HPO. Many publications, such as our publication by Heyne et al. or the recent CNV paper by Collins et al. also use HPO terminology. But in many cases, these terms are used a surrogate markers to describe conditions more broadly or identify cases that might have the condition of interest. For example, in the HPO portal, one of the clinical features of Dravet Syndrome is generalized myoclonic seizures (HP:0002123). While useful to obtain an overview of the condition, this information does not tell us how many individuals with SCN1A-related disorders have generalized myoclonic seizures and when they occur.
Atomism. The idea behind phenotypic atoms is simple. Longitudinal disease histories can be thought of as sequences of observable or measurable phenotypic events, individual data points that I would like to refer to as phenotypic atoms. These atoms have three properties: time, quality, and quantity. In the figure above, one phenotypic atom is the data point stating that this individual with an STXBP1-related disorder has daily infantile spasms at 18 months. Atoms do not have to be extremely granular with respect to temporal features, nor do they have to be very specific when it comes to describing the feature – they should simply describe observable or measurable clinical presentations as comprehensively as the clinician considers possible under the circumstances. For example, for reconstructing seizure histories, our lab uses monthly increments. We have already seen that this may not be granular enough to capture rapid changes, such as the response to ACTH in Epileptic Spasms. Neither would such a framework allow us to track seizure frequency during treatment of status epilepticus in the ICU, where important clinical events may pass by within minutes. However, this is not the primary purpose – the main goal is to have a general framework that allows us to conceptualize disease histories over months and years.
Descriptors versus atoms. The difference between simple clinical descriptions and phenotypic atoms is crucial, even though it may not always be obvious. For example, it may seem like a subtle difference whether a database lists that an individual has absence seizures or whether the same individual has daily absence seizures between 2-4 years. The distinction is that the former is a clinical descriptor, the latter is a phenotypic atom. And the phenotypic atom can easily be combined and integrated with many other longitudinal datasets. The simple descriptor cannot be easily integrated, as it lacks the necessary temporal dimensionality. For many of our studies, we have to rely on simple descriptors, but we hope that this can be increasingly replaced by longitudinal data that capture all three dimensions.
Why all this? What am I trying to accomplish by introducing a new concept about clinical data? In brief, I would like to advocate for mapping phenotypic atoms in addition to primarily thinking about outcomes and drug response, partly because analysis of these phenotypic atoms will help to prioritize disorder-specific outcome measures. The sequence of phenotypic data points constitutes the longitudinal disease history of an individual, potentially across multiple clinical dimensions rather than according to a single pre-defined outcome (atoms may relate to seizures, cognitive and psychosocial development, motor function, etc.). The collective disease histories across individuals with a shared condition represents the natural history. Many concepts of clinical endpoints implicitly rely on information provided by phenotypic atoms. Outcomes are seizures frequencies and developmental trajectories over time, while drug response represents changes in seizure frequency in response to medication exposure. Syndromes constitute constellations of phenotypic atoms that occur in specific patterns, as we had explored in Figure 4 by Fitzgerald et al., 2021. I suggest not only capturing endpoints and outcomes, but also the raw data behind this information as granular phenotypic data points including time, quality, and quantity. Clinical data are valuable, and collection of phenotypic atoms maximizes the potential for re-use: data can be re-interrogated, harmonized, and combined to test or develop clinically driven hypotheses.
What you need to know. In this blog post, I have tried to explore the underlying concept that our lab uses longitudinal phenotypic data in genetic epilepsies and neurodevelopmental disorders. In contrast to purpose-driven phenotype assessments that focus on drug response or primary outcomes, I have tried to advocate for the creation of non-purpose-driven, impassionate phenotypic atoms, collections of clinical descriptions that capture age, the type of observable or measurable phenotypic events, and their quality. Rebuilding and analyzing clinical trajectories as sequence of these interoperable data points will allow us to start making phenotypic discoveries that parallel the genomic next generation sequencing era, facilitating a more diverse perspective of the clinical manifestations of disease and their evolution that informs the development of precision medicine.
Context. I have borrowed the concept of atomism from Bertrand Russell’s Philosophy of Logical Atomism. For a good and readable overview of this short-lived period of early 20th century philosophy, I recommend The Murder of Professor Schlick by David Edmonds. This book has inspired this blog post and the idea of contrasting machine learning mysticism against the phenotypic atomism that drives our current analyses.


