SCN1A. This is the Epilepsiome page for SCN1A, one of the most well-documented and well-researched epilepsy genes. Pathogenic variants in SCN1A cause a spectrum of mild to severe epilepsy, most notably Dravet Syndrome and the mild familial epilepsy syndrome of Genetic/Generalized Epilepsy with Febrile Seizures Plus (GEFS+).
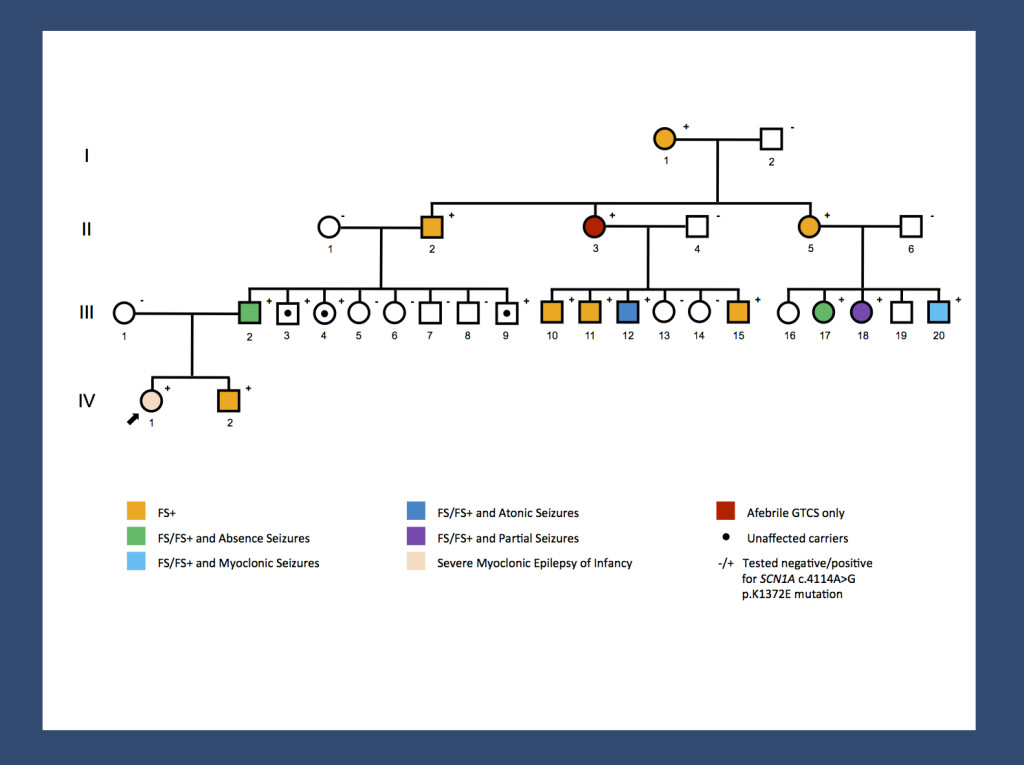
Re-posting one of our figures from November 2013 and May 2014 – a pedigree of a large GEFS+ family with 14 affected and 3 unaffected carriers published by us last year. A missense mutation results in a broad range of phenotypes spanning unaffected carriers, Febrile Seizures Plus (FS+) phenotypes and Dravet Syndrome. The range of phenotypes seen in this family suggests that SCN1A is not the entire story. It will be interesting to investigate whether the modifying factors are genetic or non-genetic. The pattern of phenotypes suggests some intra-familial factors.
Here are the most recent blog posts on SCN1A
- SCN1A – this is what you should know in 2015
- SCN1A and Dravet Syndrome – your questions for the Channelopathist
- Cause or coincidence – recessive SCN1A variants in Dravet Syndrome
- SCN1A – This is what you need to know in 2014
- From unaffected to Dravet Syndrome – extreme SCN1A phenotypes in a large GEFS+ family
In a nutshell. SCN1A is the major gene for Dravet Syndrome, a severe epilepsy of childhood most prominently characterized by fever-induced seizures. SCN1A variants can result in a wide spectrum of epilepsy phenotypes, ranging from other severe epilepsies, such as intractable childhood epilepsy with generalized tonic-clonic seizures (ICE-GTC), to milder epilepsy disorders, including simple febrile seizures or familial fever-related epilepsies referred to as Generalized/Genetic Epilepsies with Febrile Seizures Plus. More recent studies have also indicated that common variants in the SCN1A gene may be risk factors for a wide range of common epilepsies, including Temporal Lobe Epilepsy and Idiopathic/Genetic Generalized Epilepsy (GGE/IGE).
| Phenotypes | Genetics | Mechanism & Function |
| The Clinical Perspective | Community | Resources & References |
Phenotypes
Dravet Syndrome. Pathogenic variants in SCN1A are responsible for one of the most common and well-defined epileptic encephalopathies, Dravet Syndrome. Dravet Syndrome is a severe childhood epilepsy with prominent fever-associated seizures. The disease progresses to include other seizure types (myoclonic, partial), and is also associated with progressive cognitive and behavioral deficits. The population frequency of Dravet Syndrome due to variants in SCN1A is 1 in 22,000, which is comparable to that of Rett Syndrome. Dravet Syndrome is less common than other genetic disorders, but is one of the most common epileptic encephalopathies caused by a single gene.
GEFS+. Besides Dravet, SCN1A is implicated in severe fever-related epilepsies and rare familial epilepsies. Genetic Epilepsy with Febrile Seizures Plus (GEFS+) is a milder autosomal dominant familial epilepsy syndrome. The GEFS+ phenotypes span a broad range from unaffected carriers to simple febrile seizures, febrile seizures plus, and sometimes more severe epilepsies. The core phenotype in GEFS+ families are Febrile Seizures Plus. This epilepsy syndrome refers to febrile seizures (FS) persisting beyond the age of 6 years or FS in conjunction with other, afebrile seizure types. Simple FS is a very common phenotype within GEFS+ families, but the overall family generally does include members with FS+. In rare cases, GEFS+ phenotypes have been found to overlap with Dravet Syndrome within a family. The phenotypes of SCN1A in GEFS+ are obviously variable, but the source of this variability is currently unknown.
Febrile Seizures. Outside of a familial syndrome, SCN1A has been found to play a part as a susceptibility gene for Febrile Seizures. A large and well-powered study found that common variants in SCN1A predisposed a patient to Febrile Seizures.
Other Features. Individuals with pathogenic SCN1A variants often have seizures induced by environmental stimuli such as sleep deprivation. In addition, patients often have behavioral issues, including hyperactivity, impulsiveness, and distractibility. Patients with more severe epilepsy phenotypes can develop postural changes and ataxia.
Other Phenotypes. Two association studies have suggested common variants in SCN1A as risk factors for Idiopathic/Genetic Generalized Epilepsies (IGE/GGE) and Temporal Lobe Epilepsy. Now, a recent study in Brain finds an intronic variant of the SCN1A gene that is associated with Temporal Lobe Epilepsy (TLE), the most common epilepsy in man. Interestingly, the association with SCN1A seems to be specific for only a particular subtype of focal epilepsies.
De novo mutations in SCN1A are occasionally identified in other severe childhood epilepsies including Myoclonic Astatic Epilepsy (MAE), Lennox-Gastaut Syndrome, and Migrating Partial Epilepsy of Infancy (MMPSI). These findings, however, are too rare to produce a coherent picture of additional SCN1A-related epilepsies. The take-home message is that other phenotypes can also be the consequence of an underlying SCN1A mutation.
SCN1A variants have also been identified in families with familial hemiplegic migraine with or without epilepsy.
Genetics
Dravet. Dravet Syndrome is caused by de novo alterations in the SCN1A gene in more than 80% of patients. Even in patients with reportedly negative SCN1A testing, some patients may have SCN1A variant at second glance.
SCN1A more common. Various studies using next-generation sequencing find “hidden” SCN1A variants also in patients that were previously considered SCN1A negative. This observation suggests that the Dravet Syndrome phenotype is tightly linked to SCN1A and that a negative SCN1A result in a patient with classic Dravet Syndrome is probably more likely to be a false negative than due to a different gene. To put this differently: if you have a patient with typical Dravet Syndrome and a negative test for SCN1A with traditional Sanger sequencing, you are probably more likely to find a “hidden” SCN1A variant on re-testing than to find a variant in a different gene. Data is currently coming together, which will reveal whether this relatively bold assumption is true.
Mutation Spectrum. Over 1,250 different disease-causing variants have been reported throughout SCN1A. Most variants are heterozygous missense or nonsense variants. Splice site variants, small insertions or deletions, and large exonic or whole gene deletions are also relatively common. A small number of exonic or whole gene duplications and complex rearrangements have also been identified. Most pathogenic variants in SCN1A occur in the C-terminus and in the pore loops connecting S5 and S6. Many SCN1A variants are unique to a single patient or a small number of patients.
Databases. The SCN1A gene is not only a well-researched, but also a well-documented gene. There are at least two databases in addition to ClinVar and HGMD that review and interpret the reported mutations in the SCN1A gene including the Antwerp SCN1A database and a recently published database that was last updated in February 2015. Accordingly, there is much data out there regarding the mutational spectrum about this gene. It will be interesting to see whether any new patterns will emerge from these large datasets.
Genotype-Phenotype Correlation. There is some correlation between the type of the mutation and the resulting phenotype. For example, variants that truncate or completely disrupt the SCN1A protein are more likely to be found in patients with Dravet Syndrome than in patients with GEFS+, while missense variants are more common in patients with GEFS+. In addition, de novo variants are more common in patients with a more severe phenotype, such as Dravet syndrome, while inherited variants are more frequent in patients with GEFS+. However, prediction of the phenotype based on a specific variant or variant type is not possible, as patients with the same variants can have a broad range of phenotypes. SCN1A testing remains diagnostic, not predictive.
General Considerations for Variant interpretation. When reviewing a genetic variant to determine its significance for a given patient, it is important to weigh multiple pieces of evidence:
Considerations for gene level interpretation. First, it is important to establish the strength of the evidence showing that the gene is associated with epilepsy. Some genes may only have one variant reported in a single individual with epilepsy, while other genes may have multiple variants reported in many large families with an autosomal dominant pattern of epilepsy. For SCN1A, there is very strong evidence for its role in human epilepsy.
Considerations for variant level interpretation. When reviewing the significance of a variant, it is important to consider the impact on the gene and the presence of the variant in previously described patient and control populations. Many clinical genetic testing laboratories classify genetic variants into different categories, ranging from benign to pathogenic. Variants that are common in control populations and would not be predicted to have a major impact on the gene/protein are generally classified as benign. Variants are more likely to be classified as pathogenic if the variants are rare or not present in the control population, reported in multiple individuals or families with disease, and likely to have a higher impact on the gene/protein based on the type of mutation or functional studies. Variants with uncertain or limited available evidence may be classified as variants of uncertain significance (VUS), indicating that further information is required in order for the variant to be further defined. In some cases, testing additional family members can be helpful, as it allows the lab to determine whether or not the variant was inherited (versus de novo) and how the variant segregates with disease in the family. Sometimes further classification of a VUS requires waiting for the identification of additional patients or families with similar or nearby variants.
Inheritance, Penetrance & Prevalence. SCN1A-related epilepsies are inherited in an autosomal dominant pattern. Most patients with a more severe SCN1A-related phenotype have a de novo variant. SCN1A-related disorders can have reduced penetrance and significant variable expressivity, even with the same variant and within the same family, The exact prevalence of SCN1A-related epilepsy is unknown.
Recessive Inheritance? Some publications claim to have identified families and individual patients with recessive inheritance in SCN1A. These findings require some caution in interpretation since, particularly with recessive variants, it is difficult to tell signal from noise.
Modifier genes not ready for prime time. There are various studies on modifier genes of SCN1A, suggesting that variants in other genes may modify the phenotype in patients with pathogenic SCN1A variants. In a nutshell, none of these studies is sufficiently replicated to securely implicate a modifier gene. Sufficiently powered studies, however, will hopefully be possible in the near future with increasing numbers of patients.
Mechanism & Function
Sodium Channels. SCN1A encodes for the alpha-1 subunit of the voltage-gated sodium channel (Na(V)1.1). The transmembrane alpha subunit forms the central pore of the channel. This ion channel is critical to the generation and propagation of action potentials. The channel responds to the voltage difference across the cell membrane to create a pore that allows sodium ions through the membrane. The influx of sodium creates an action potential, which is critical to signaling within the brain. In some animal models, loss-of-function mutations were shown to lead to a reduction in sodium currents and impaired signalling of GABAergic interneurons of the hippocampus.
In 70-90% of cases, Dravet Syndrome is caused by a de novo mutation in SCN1A, which often leads to a non-functional protein and possibly results in haploinsufficiency. Haploinsufficiency refers to the situation when only a single functional copy of a gene is present, and the gene product of only one copy is insufficient for normal functioning. However, some SCN1A variants seem to result in a gain-of-function. Therefore, as is the case with many other epilepsies, we are still not sure of the exact mechanism that leads to SCN1A epilepsies. SCN1A mutations that result in GEFS+ exist in many different parts of the gene and can result in various functional effects on the SCN1A protein, predicted to cause either hypo- or hyperexcitability of the neuron.
Cell/Animal models.
Mice. The SCN1A knockout mouse has helped to elucidate the underlying disease mechanism by showing that the mice have altered function of Nav1.1 sodium channels in neurons throughout the brain. Studies also suggest that impaired γ–aminobutyric acid (GABA)ergic firing in hippocampal interneurons results in seizure susceptibility and impairment of the GABAergic cerebellar Purkinje cells leads to motor issues, such as ataxia.
Studies of SCN1A mouse models have also helped to elucidate the high risk of SUDEP in individuals with SCN1A variants. The mice seem to have increased parasympathetic activity following tonic– clonic seizures, resulting in lethal bradycardia and electrical ventricular dysfunction. Another study showed potential predisposition to cardiac arrhythmia, which may also contribute to the risk of SUDEP.
Zebrafish. Zebrafish models of SCN1A-related disorders are also helping to further characterize the underlying disease pathogenesis and response to variant pharmacological agents.
The Clinical Perspective
Recurrence risk & testing of family members. SCN1A variants can be de novo or inherited in an autosomal dominant pattern. Each child of an individual with a disease-causing variant has a 50% (1 in 2) of inheriting the disease-causing variant. Somatic and germline mosaicism of SCN1A variants have been reported, making the recurrence risk for families with a child with an apparent de novo variant higher than the general population risk.
Therapy. Seizures in patients with pathogenic SCN1A variants respond most optimally to antiepileptic drugs that bind the GABA receptor. Good seizure control is essential because it can promote better cognitive function, even in patients who have shown decline and drug resistance over an extended period of time, and decrease the risk of injury and sudden unexplained death in epilepsy. Studies suggest that the encephalopathy in patients with Dravet syndrome is not purely caused by the associated epilepsy but at least partially a consequence of the underlying channelopathy. Wallace and colleagues provide a nice review of therapy options for Dravet syndrome.
A number of anti-epileptic drugs are contraindicated due to side effects: carbamazepine, lamotrigine and vigabatrin can cause myoclonic seizures, while phenytoin can cause choreoathetosis. In addition, seizure triggers should be avoided when possible, including hot temperatures (warm baths, exercise on hot days, untreated fever) and photosensitivity triggers.
Community
Research studies. The scientific community is actively studying SCN1A and its role in human disease. The SCN1A Epilepsiome team is happy to facilitate if you have questions or a specific interest in this gene.
Family connections. The Dravet Syndrome Foundation provides support and information for families dealing with a Dravet diagnosis. The ICE Epilepsy Alliance focuses on providing resources for children affected by intractable epilepsies, particularly ion channel epilepsies.
Patient registry. The epilepsy community has a registry to gather information about individuals with SCN1A variants or variants in other genes involved in ion channel function: International Ion Channel Epilepsy Patient Registry.
Resources & References
Websites
For Families:
For Clinicians: SCN1A-Related Seizure Disorders GeneReviews
References
Dravet syndrome–from epileptic encephalopathy to channelopathy.
Brunklaus A, Zuberi SM.
Epilepsia. 2014 Jul;55(7):979-84.
PMID: 24836964
First mutation in the voltage-gated Nav1.1 subunit gene SCN1A with co-occurring familial hemiplegic migraine and epilepsy.
Castro MJ, Stam AH, Lemos C, de Vries B, Vanmolkot KR, Barros J, Terwindt GM, Frants RR, Sequeiros J, Ferrari MD, Pereira-Monteiro JM, van den Maagdenberg AM.
Cephalalgia. 2009 Mar;29(3):308-13.
PMID: 19220312
Five-year extended follow-up status of 10 patients with Dravet syndrome treated with fenfluramine.
Ceulemans B, Schoonjans AS, Marchau F, Paelinck BP, Lagae L.
Epilepsia. 2016 May 20.
PMID: 27197941
De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy.
Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P.
Am J Hum Genet. 2001 Jun;68(6):1327-32.
PMID: 11359211
Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2.
Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, Buresi C, Malafosse A.
Nat Genet. 2000 Apr;24(4):343-5.
PMID: 10742094
Early-onset familial hemiplegic migraine due to a novel SCN1A mutation.
Fan C, Wolking S, Lehmann-Horn F, Hedrich UB, Freilinger T, Lerche H, Borck G, Kubisch C, Jurkat-Rott K.
Cephalalgia. 2016 Jan 12.
PMID: 26763045
Mutations of sodium channel alpha subunit type 1 (SCN1A) in intractable childhood epilepsies with frequent generalized tonic-clonic seizures.
Fujiwara T, Sugawara T, Mazaki-Miyazaki E, Takahashi Y, Fukushima K, Watanabe M, Hara K, Morikawa T, Yagi K, Yamakawa K, Inoue Y.
Brain. 2003 Mar;126(Pt 3):531-46.
PMID: 12566275
A Point Mutation in SCN1A 5′ Genomic Region Decreases the Promoter Activity and Is Associated with Mild Epilepsy and Seizure Aggravation Induced by Antiepileptic Drug.
Gao QW, Hua LD, Wang J, Fan CX, Deng WY, Li B, Bian WJ, Shao CX, He N, Zhou P, Liao WP, Shi YW.
Mol Neurobiol. 2016 Mar 11.
PMID: 26969601
Altered Glycolysis and Mitochondrial Respiration in a Zebrafish Model of Dravet Syndrome.
Kumar MG, Rowley S, Fulton R, Dinday MT, Baraban SC, Patel M.
eNeuro. 2016 Apr 5;3(2). eCollection 2016 Mar-Apr.
PMID: 27066534
Identification of an Nav1.1 sodium channel (SCN1A) loss-of-function mutation associated with familial simple febrile seizures.
Mantegazza M, Gambardella A, Rusconi R, Schiavon E, Annesi F, Cassulini RR, Labate A, Carrideo S, Chifari R, Canevini MP, Canger R, Franceschetti S, Annesi G, Wanke E, Quattrone A.
Proc Natl Acad Sci U S A. 2005 Dec 13;102(50):18177-82.
PMID: 16326807
Low-dose fenfluramine in the treatment of neurologic disorders: experience in Dravet syndrome.
Schoonjans AS, Lagae L, Ceulemans B.
Ther Adv Neurol Disord. 2015 Nov;8(6):328-38.
PMID: 26600876
Pharmacotherapy for Dravet Syndrome.
Wallace A, Wirrell E, Kenney-Jung DL.
Paediatr Drugs. 2016 Jun;18(3):197-208.
PMID: 26966048
Sodium channel alpha1-subunit mutations in severe myoclonic epilepsy of infancy and infantile spasms.
Wallace RH, Hodgson BL, Grinton BE, Gardiner RM, Robinson R, Rodriguez-Casero V, Sadleir L, Morgan J, Harkin LA, Dibbens LM, Yamamoto T, Andermann E, Mulley JC, Berkovic SF, Scheffer IE.
Neurology. 2003 Sep 23;61(6):765-9.
PMID: 14504318

