Phenotypic bottleneck. This is another post in the “phenotypic atomism series,” what has become our lab’s philosophy in how we think about and work with longitudinal clinical data. However, before we introduce another dimension to the phenotypic atom, let me first revisit the idea of the “phenotypic bottleneck” – a concept that had piqued my interest three years ago and led me to join the lab. In brief, in contrast to established pipelines for large-scale analysis of sequencing data, our ability to analyze clinical data at scale remains more limited. As a result, phenotypic characterization lags behind gene discovery, even with tremendous progress in the last few years. A major challenge stems from the inherent nature of working with multi-dimensional longitudinal clinical data: it can be sparse and incomplete at times. However, how much of the unknown is truly unknown?
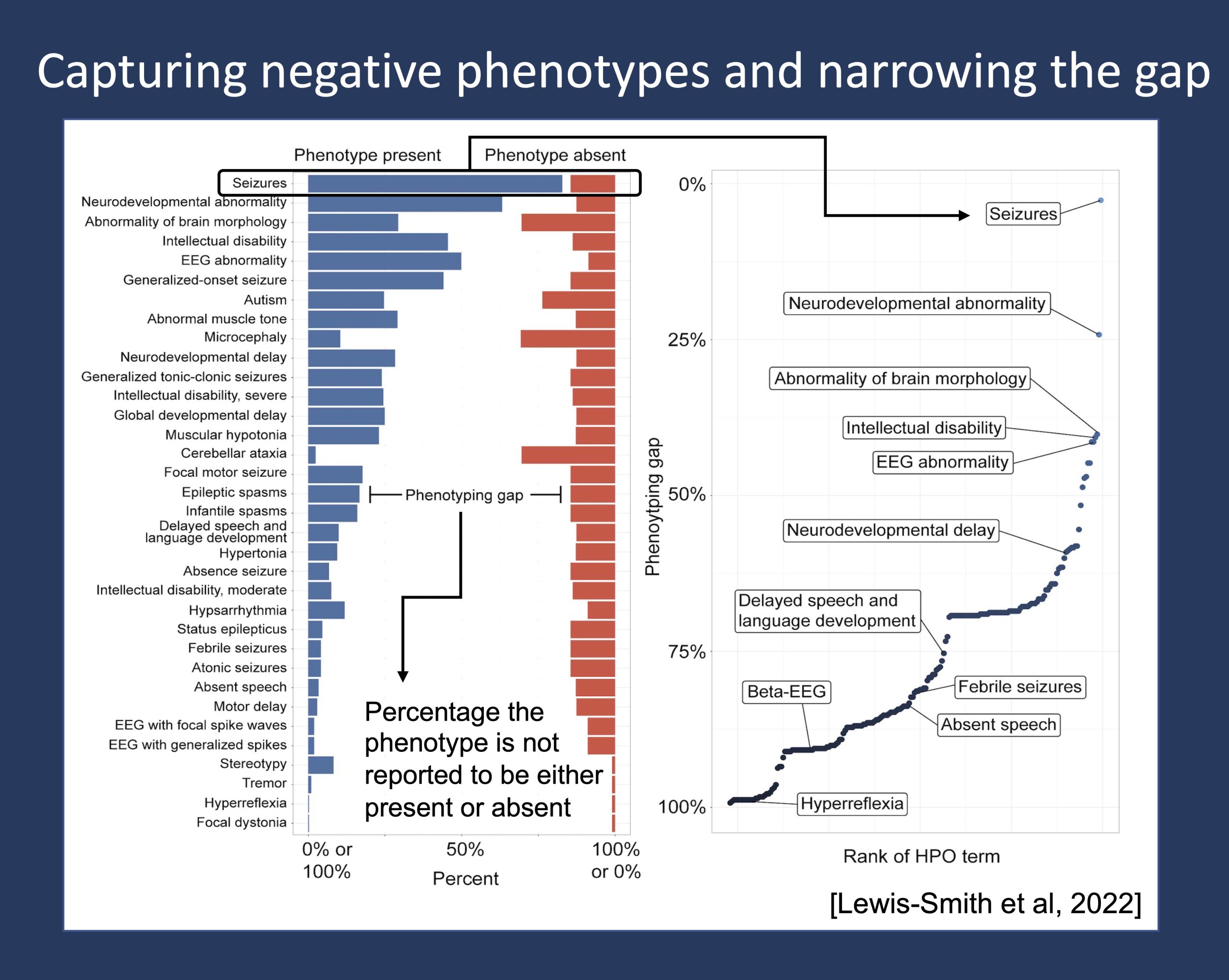
Figure 1. The phenotypic gap represents the known unknown – the percentage of the time a clinical feature is not explicitly documented to be either present or absent, i.e., not reported to be either positive or negative phenotypes. Adapted from Lewis-Smith et al, this figure shows the gap when assessing a range of clinical features in a cohort of 413 individuals with SCN2A-related disorders, a group of conditions associated with epilepsy and autism, highlighting the difference in gap in more commonly captured features such as the presence or absence of seizures compared to less commonly captured features such as hyperreflexia.
Negative phenotypes. When, for example, seizures are not reported in an individual in the Electronic Medical Records (EMR), with what degree of certainty or confidence can we say that seizures are truly absent? To answer this question, it first requires giving rise to the concept of a negative phenotype. In contrast to the presence of a clinical feature, or what we call a positive phenotype, we refer to the absence of a clinical feature as a negative phenotype. However, just because a clinical feature was not reported or documented does not mean that it takes on the meaning of a negative phenotype. We only code phenotypes as negative if they are truly known to be absent – clinical features that were specifically assessed and explicitly reported to be absent at time of assessment. When coding negative phenotypes, our own internal convention is to negate the positive phenotype version of the Human Phenotype Ontology, or HPO (e.g., ‘No seizure’ becomes notated as ‘NP:0001250’, which is the negated form of ‘Seizure’ and ‘HP:0001250’).
Harmonization. As for positive phenotypes, leveraging the standardized and relational framework of the HPO for negative phenotypes allows us to make implicit connections between features. For example, we can conclude that if an individual does not have seizures, then they also do not have either generalized-onset seizures or focal-onset seizures, or generalized myoclonic seizure or focal impaired awareness seizure, and so forth. However, you can imagine that this type of reasoning can generate a massive dataset that includes significant redundancy and requires a different way of thinking; we have started to develop the framework for integrating negative phenotypes, including how we only capture the information that holds additional value. But why is this important, and what is the point?
Atomic dimensionality. While we typically talk about the presence of a phenotypic feature, the understanding that a phenotypic feature is absent can be just as, if not more, informative. For example, individuals with KCNQ2-related disorder and self-limited neonatal-infantile seizures have early-onset seizures but typical development – an absence of neurodevelopmental abnormalities, which differentiates this cohort from a cohort of individuals with KCNQ2-related developmental and epileptic encephalopathy. There is understated value in knowing what is absent, and it is for this reason we define this component as another dimension of the phenotypic atom.
Phenotypic gap. So, to what degree are phenotypes captured in either their positive or negative states, and how much do we actually know? In our prior publication on a cohort of 413 individuals with SCN2A-related disorders, we asked this exact question (Figure 1). We know that in this cohort of individuals with neurodevelopmental disorders associated with epilepsy and autism, the most assessed phenotype is, unsurprisingly, seizures. In fact, more than 97% of the cohort were reported to either have or not have seizures. However, there remains a relatively wide gap for almost every other clinical feature. And with the median gap at 80%, there remains open discussion on how we should resolve missing data.
Widening the bottleneck. While more granular documentation may appear to be an intuitive solution, it is impractical and inherently impossible to assess for the presence or absence of every clinical feature, especially for rare phenotypes and when assessing existing data. So, when seizures are not reported in an individual, with what degree of certainty can we say that seizures are truly absent? In our SCN2A study, it can be expected and is shown accordingly in Figure 1 that the occurrence of seizures is well documented, including the absence of seizures. Thus, it can be reasonable to make the Closed World Assumption that the 3% of individuals not reported to either have or not have seizures, likely do not have seizures. However, for most other clinical features with a wider phenotypic gap, we must take to an Open World Assumption. Much is unknown and points to the importance of negative phenotypes in filling the gap and allowing us to make more precise conclusions and calculations, especially as we continue to develop conceptual frameworks and advance methods from NLP and phenotype imputation to machine learning and modeling.
What you need to know. When talking about capturing longitudinal clinical data, a dimension that has been and still remains overlooked is the absence of clinical features. And what I mean by absence is not the absence of knowledge in whether we know a phenotype to be present but instead, the explicit absence of a clinical presentation. As previously suggested, we may actually know more than we think – with the wealth of data and thousands of patient years of clinical histories captured from ongoing care in the EMR, there stands to be untapped value in differentiating and providing higher resolution to the clinical features that we ultimately measure.


