GRIN2A is a gene with wide phenotypic variability—often within the same family. GRIN2A encodes for a subunit of the NMDA receptor and pathogenic variants have been linked to focal epilepsy, speech disorders, and neurodevelopmental differences.
Here are the most recent blog posts on GRIN2A
- GRIN2A – this is what you need to know in 2015
- Speech dyspraxia and dysarthria – the other side of GRIN2A
- GRIN2A encephalopathy, epilepsy-aphasia and rolandic spikes
- Publications of the week: GRIN2A, SCN8A, and DEPDC5
- EuroEPINOMICS and the golden age of epilepsy gene discovery
In a nutshell. GRIN2A is associated with a wide spectrum of phenotypes, and can have variable presentations even without the same family. However, prominent features include learning differences including intellectual disability, specific learning disabilities, and speech disorders, as well as focal/multifocal epilepsy and epilepsy aphasia. While genotype-phenotype correlations are incomplete, it is expected that variants leading to a developmental and epileptic encephalopathy phenotype are gain of function.
Phenotype | Genotype | Mechanism | Community
Phenotype
Phenotypes. The spectrum of phenotypes, as we understand them, associated with genetic alterations in GRIN2A is very broad and ranges from possibly unaffected to severe encephalopathy.
Learning and speech. Mild intellectual disability (ID) is a common feature in individuals carrying a pathogenic variant in GRIN2A—though there has been a range of severity reported. ID is believed to occur independently of speech and epilepsy. In some families, affected individuals present with a relatively mild phenotype of learning disability and/or mild speech disorders – not necessarily associated with epileptic seizures.
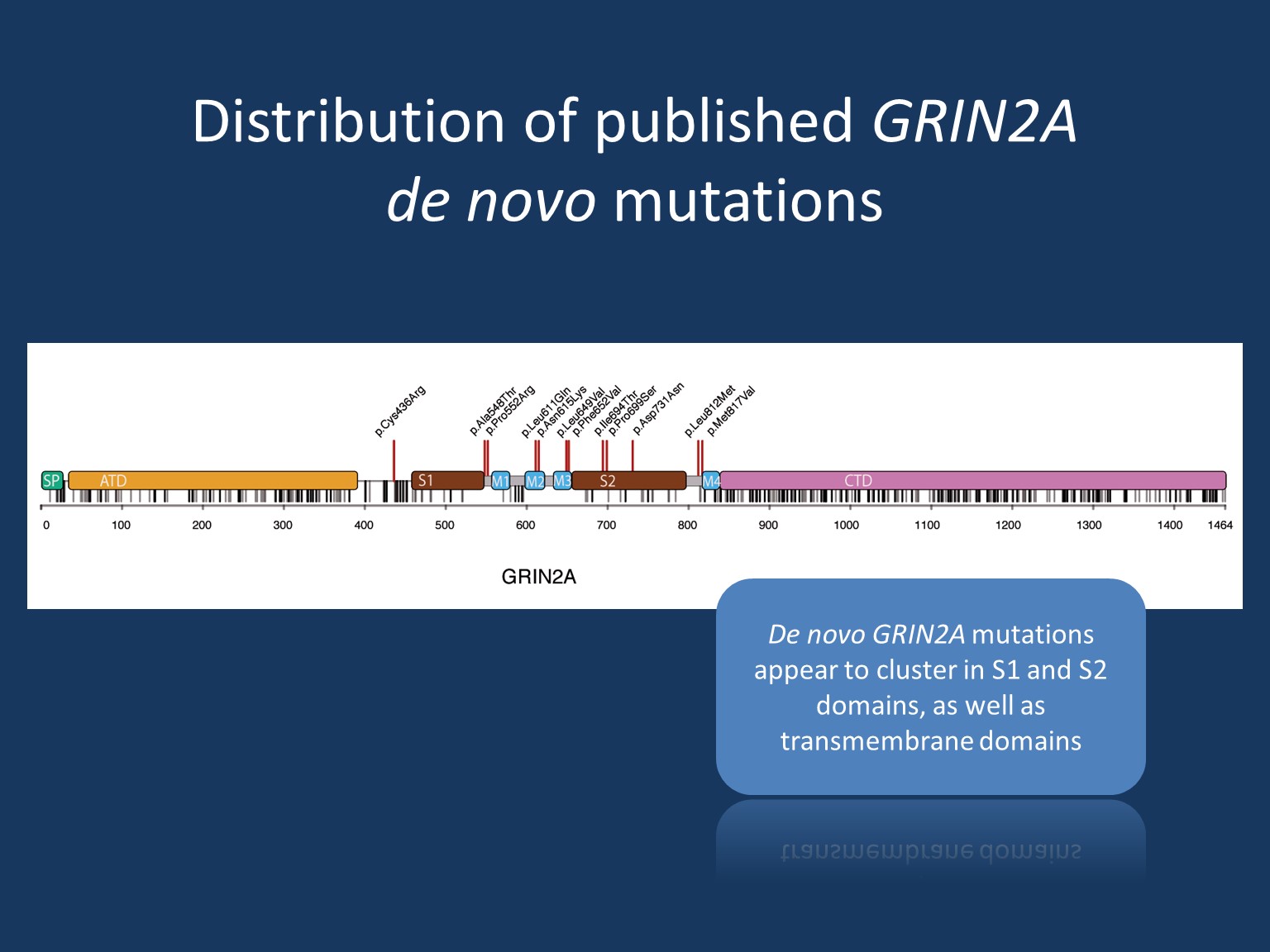
Epilepsy. Epilepsy in GRIN2A includes spectrums of focal/multifocal epilepsy and epilepsy aphasia. The spectrum of focal epilepsy with centrotemporal spikes comprises classic Rolandic epilepsy (RE), atypical benign partial epilepsy (ABPE), continuous spikes and waves during slow-wave sleep syndrome (CSWS) and Landau-Kleffner syndrome (LKS). The epilepsy aphasia shows phenotypic overlap in particular to CSWS and LKS. Finally, severe epileptic encephalopathy is a rarer but known presentation of GRIN2A, particularly in those with de novo missense variants in the transmembrane or linker domains.
Unaffected individuals. Patients may inherit a pathogenic GRIN2A variant from an apparently unaffected parent, though it currently remains unclear whether these carriers have subclinical phenotypes (such as EEG abnormalities in childhood). Given this, some have suggested that GRIN2A may have reduced penetrance, though this remains speculative until the clinical features of supposed unaffected carriers are further characterized.
Genotype
There are still many questions surrounding genotype-phenotype correlates in GRIN2A. At this time, it is believed that pathogenic missense variants located in the transmembrane or linker domains are associated with severe developmental and epileptic encephalopathy. In contrast, pathogenic missense variants in the amino-terminal or ligand-binding domains have been reported to have wider phenotypic variability. Phenotypes described include ID within a spectrum of severity—though more often mild—as well as focal epilepsy and/or speech disorders. Individuals carrying truncating variants in GRIN2A appear not to cause epilepsy encephalopathy but rather the range of other phenotypes described in the prior section.
Mechanism
GRIN2A encodes the glutamate-binding GluN2A subunit of the NMDA receptor in the excitatory synapsis. The assembly of this heterotetrameric receptor follows a specific temporo-spatial expression pattern.
Loss-of-function. Given the phenotypic similarity, it is believed that truncating variants and pathogenic missense variants located in the amino-terminal and ligand-binding domains have a similar mechanism. In turn, pathogenic missense variants in the transmembrane or linker domains may operate differently on a molecular level. It was previously hypothesized that a lack of GluN2A expression due to truncation and haploinsufficiency would subsequently be compensated by an increase of GluN2B expression, however, recent evidence suggests that this compensatory mechanism does not hold true in physiological models. Rather, haploinsufficiency is now the presumed mechanism for truncating variants as well as variants located in the amino-terminal/ligand-binding domains.
Gain-of-function. Considering the stark contrast in phenotype, it is unlikely that pathogenic missense variants in the transmembrane/linker domains have the same mechanism as the aforementioned. Rather, it is likely that these variants have a form of a gain-of-function mechanism. When considering the mechanistic possibilities of gain-of-function, one must consider a resulting dominant-negative effect vs. an effect leading to a unique and abnormal functional outcome. In the case of a dominant-negative effect, one can suspect a loss-of-function >50% but not to the extent of a total loss. Researchers may gain insight into the nature of the gain-of-function effects related to GRIN2A through memantine, a specific NMDA receptor blocker that has been shown to normalize NMDA receptor function in vitro. Application of memantine in a patient with GRIN2A encephalopathy resulted in marked reduction of seizure frequency and other improvements—suggesting the mechanism of pathogenic variation to be related to functional overactivity. However, this observation on a single patient remains to be confirmed in additional cases. Conversely, a carrier of compound heterozygous truncating variants in GRIN2A was reported with severe developmental and epileptic encephalopathy. This report provides unique insight into the phenotype of a total loss. This perhaps suggests that a dominant-negative mechanism remains a possible cause of GRIN2A epileptic encephalopathy.
Variability. At this time, it remains unclear what additional factors might modify phenotypic severity—particularly for family members carrying identical GRIN2A variants. Given that phenotypic variability is most likely associated with haploinsufficiency, one may presume that each carrier is predisposed to a variety of factors that may/may not increase compensatory mechanisms for the associated loss.
Community
References
Chen W, Tankovic A, Burger PB, Kusumoto H, Traynelis SF, Yuan H. Functional evaluation of a de novo GRIN2A mutation identified in a patient with profound global developmental delay and refractory epilepsy. Mol Pharmacol 2017; 91: 317–30.
Pierson TM, Yuan H, Marsh ED, Fuentes-Fajardo K, Adams DR, Markello T, Golas G, Simeonov DR, Holloman C, Tankovic A, Karamchandani MM, Schreiber JM, Mullikin JC; PhD for the NISC Comparative Sequencing Program; Tifft CJ, Toro C, Boerkoel CF, Traynelis SF, Gahl WA. GRIN2A mutation and early-onset epileptic encephalopathy: personalized therapy with memantine. Ann Clin Transl Neurol. 2014 Mar 1;1(3):190-198.
Strehlow V, Heyne HO, Vlaskamp DRM, Marwick KFM, Rudolf G, de Bellescize J, Biskup S, Brilstra EH, Brouwer OF, Callenbach PMC, Hentschel J, Hirsch E, Kind PC, Mignot C, Platzer K, Rump P, Skehel PA, Wyllie DJA, Hardingham GE, van Ravenswaaij-Arts CMA, Lesca G, Lemke JR; GRIN2A study group. GRIN2A-related disorders: genotype and functional consequence predict phenotype. Brain. 2019 Jan 1;142(1):80-92.
Strehlow V, Rieubland C, Gallati S, Kim S, Myers SJ, Peterson V, Ramsey AJ, Teuscher DD, Traynelis SF, Lemke JR. Compound-heterozygous GRIN2A null variants associated with severe developmental and epileptic encephalopathy. Epilepsia. 2022 Oct;63(10):e132-e137.
Swanger SA, Chen W, Wells G, Burger PB, Tankovic A, Bhattacharya S et al.. Mechanistic insight into NMDA receptor dysregulation by rare variants in the GluN2A and GluN2B agonist binding domains. Am J Hum Genet 2016; 99: 1261–80.