FIRES. Febrile infection-related epilepsy syndrome (FIRES) is characterized by refractory status epilepticus following a non-specific febrile illness. FIRES is a subtype of New Onset Refractory Status Epilepticus (NORSE) without a clear cause in individuals without active epilepsy. The cause of FIRES and NORSE is unclear, and it is not even clear whether both conditions share a joint mechanism or represent distinct entities. In a recent publication, we contributed to a review of the state-of-the-art in NORSE and FIRES research and suggested a very first step to understand these conditions better – standardized biosamples. This blog post is about the intersection of omics and urgency, long-term strategies and scientific principles.
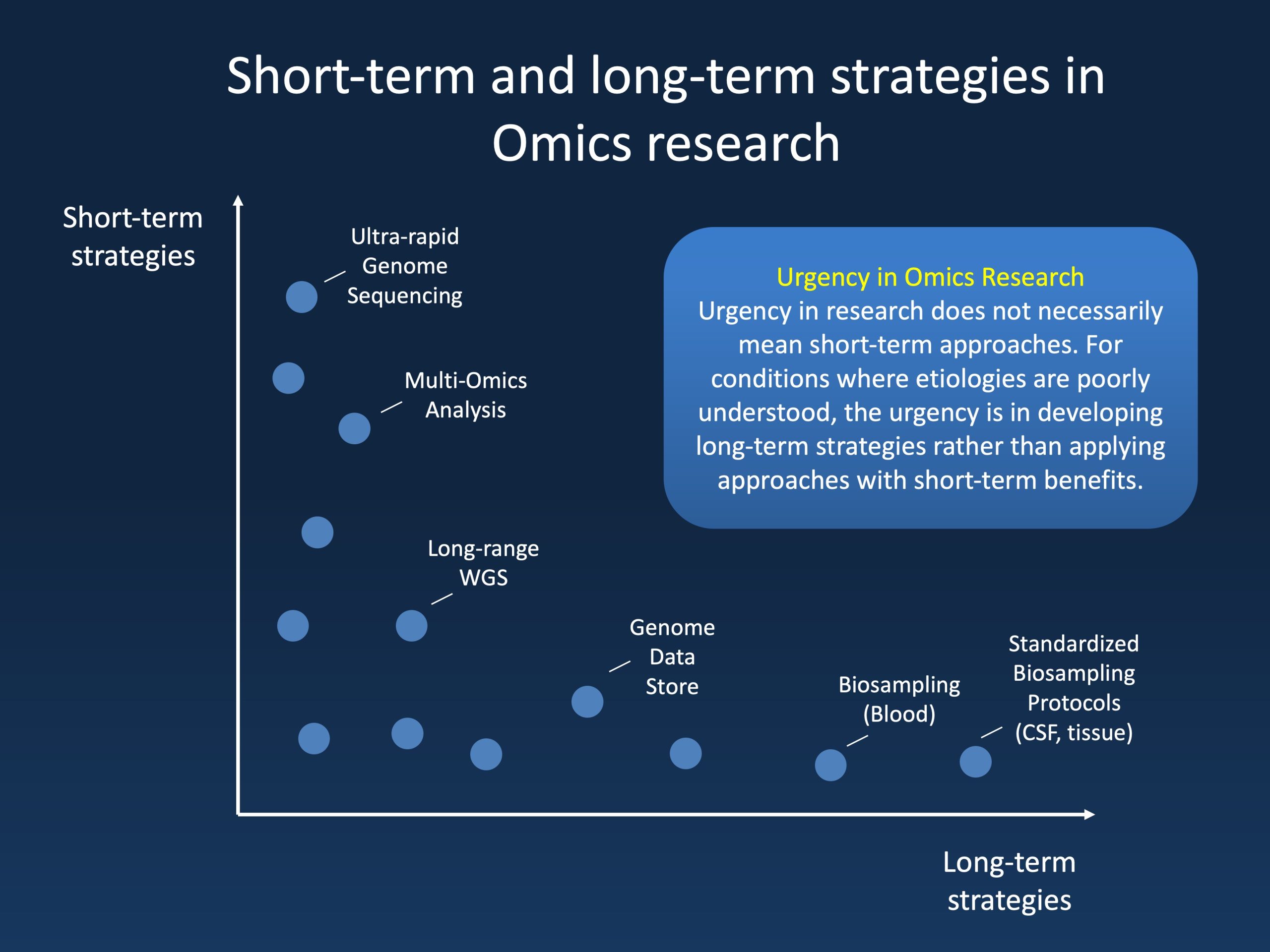
Figure 1. Depending on the condition to be studied, urgency may involve short-term or long-term approaches. For well-established conditions, an increase in the speed and application of new modalities such as rapid genome sequencing and multi-omics analysis are relevant. In contrast, in conditions with poorly understood etiologies, long-term strategies to enable future discoveries may be more impactful. The chart above lists short-term and long-term approaches on a hypothetical spectrum.
Urgency. A few months ago, I found myself on a somewhat interesting side of a scientific debate. The topic to be discussed was the plan for a specific rare (non-neurological) condition that was to be better understood through multi-omics approaches – proteomics, metabolomics, transcriptomics in addition to whole genome sequencing (WGS). The team involved argued to use as many technologies as rapidly as possible given the urgency of the condition, and their sense of purpose and dedication to their patients was palpable in the room.
Debate. The temperature in the room then dropped to zero when I made the statement “I don’t think this strategy makes sense – without these tools being more established, you will likely have too much variation to see any signal – focus on fewer technologies with larger sample sizes.” The debate went back and forth a few times: urgency versus omics humility. We eventually agreed on not disagreeing, but I left the discussion feeling like a the odd one out. Why would I suggest not doing everything as quickly as possible to potentially gain any insight that may help treatment, especially when (as in this case) funding is not an issue?
Reasons. There are two main reasons for my reaction that day. First, I draw a very strict line between clinical care and research. Clinical care is urgent, and families have a right towards getting the care they need as quickly as needed. Research, in contrast, is about creating generalizable knowledge. The urgency is not primarily in treating individuals but in generating a knowledge framework to make such treatments possible in the first place. For example, without research, there would not have been a discovery of Glut1 or SCN1A, findings that are critical for planning better treatments. Second, not every application of omics technology may be scientific in the narrow sense. In some cases, the omics approach of “finding anything without a prior hypothesis” can be in direct contrast to the scientific method. A scientific theory should be one thing above everything else: falsifiable. And which theory do we allow to be falsified by a multi-omics analysis in a small cohort?
FIRES/NORSE. This thought brings me back to FIRES and NORSE. In the recent publication by Hanin and collaborators, we reviewed the current state of biomarker research in FIRES and NORSE, looking at a comparable scenario. Both conditions are so severe that it often feels that there must be something around the corner. For a few years, we thought that if we only had genome sequencing, we would be able to point at a genetic cause. In parallel, many other groups had related thoughts in their domain. This has resulted in a situation where most studies looking at biomarkers in FIRES and NORSE were performed on small patient cohorts and lacked control patients, making it virtually impossible to validate these finding. Speaking about genomics, our own field, the genetic yield for monogenic etiologies in FIRES is zero. Yet, we would never feel that genome sequencing in FIRES would not be appropriate for this – the clinical urgency is too high to miss a potentially treatable condition even though we never identified one. The same is true to autoimmune markers. Despite publications hinting at the role of autoimmune/inflammatory mechanisms, there is little direct evidence that FIRES is an autoimmune condition or inflammatory condition more broadly – yet we would always assess individuals for this given the severity of the condition and the potential benefits if a cause was to be found.
Biosampling. So, what should we do scientifically to get closer to understanding the mechanisms underlying FIRES and NORSE? The answer may sound counter-intuitive. The next logical step is for improving and aligning standardized biosampling. Only with sufficient sample numbers ascertained and processed in the same way will it be possible to systematically study these conditions. While this may feel like going in the wrong direction, there are many examples for this approach in other disorders, for example, pediatric brain cancer. In summary, for FIRES and NORSE, it might potentially be worthwhile to leave omics tools aside for the time being and focus on biosample acquisition. This does not mean that clinical omics should not be performed – quite the opposite. It is just important to realize that these will be clinical questions directly aiming at patient care rather than research questions.
What you need to know. Omics and urgency are two different concepts that potentially misalign. For FIRES and NORSE, two conditions that are often studied in limited cohorts due to the rarity of these conditions, there is tremendous urgency to understand these conditions better. However, with limited resources, the urgency is not in applying omics technologies at scale. In contrast, the urgency is to create a framework that will enable such discoveries through establishing standardized biosampling protocols to ensure that biomaterial will be available in the first place for multi-omics approaches.


