The backbone. As we have started a new round for BENCH introductory sessions with new collaborators, I thought that it might be timely to talk a little bit about our BENCH phenotype database and the concepts behind it. In addition to the purely technical aspects, there is a more fundamental question behind this: how do we want to document and store epilepsy phenotypes for research purposes, how do we find the balance between precision and efficiency?
About EuroEPINOMICS. When we initially thought about the structure of the EuroEPINOMICS Collaborative Research Project on Rare Epilepsy Syndromes (EuroEPINOMICS-RES), one thing became quite clear from the very beginning: even though RES would be working in the format of a large genetic collaborative project, there would be no centralized money for institutions like a biobank or phenotype database. EuroEPINOMICS is part of the EUROCORES scheme of the European Science Foundation. This means that the entire program consists of individual subprojects that are supported by the national funding agencies. For us in Germany, this is the German Research Foundation (DFG) and our grant is the same as a regular project grant. And biobanks and phenotype databases are infrastructure, not actual research projects. How did we try to solve this problem? We tried to build a phenotype database that is a research project in itself. This is how we came up with BENCH-RES.
Birds and feathers. In evolutionary biology, there is a concept called exaptation. A function of a trait might shift during its evolutionary history, as in the example of feathers. Feathers evolved in the context of thermoregulation and once feathers were evolved, it turned out that they can also be used for flying. Something similar occurred with our database. For our genetic and phenotypic data, we decided to use the BENCH database by Cartagenia. BENCH can be used to store information on array-CGH and SNP-array for genetic laboratories and research groups. In order to handle the phenotypic data, BENCH allows for a structured data entry and an ontology-based data entry. However, we currently store very little genetic data in the BENCH database, as we “exapted” the database system as a phenotype database. It turned out that the latter has become our main focus when using BENCH and that particularly the ontology-based phenotype data is very interesting. But what exactly are ontologies?
Phenotype, phenotype. Assessing and selecting phenotypic information for research purposes is sometimes difficult, as we have to find the balance between sufficient detail and efficiency. Assessing the complete epilepsy phenotype with several hundred variables might give us a good idea about the patient’s phenotype, but we might run into problems when trying to do this on larger scale. This is simply a matter of time and manpower. In addition, some of the detailed data might not be very informative. Which brings us to the concept of “information content”. Using our general understanding of information, probably everybody would agree that a more specific phenotypic term is more informative than a more general term. For example, “acoustic aura” is more informative than “focal seizure”, “absence seizure with perioral myoclonias” is more informative than “dialeptic seizure”. The more specific terms are more informative not because they are necessarily more detailed, but because they delineate a smaller patient subgroup. For example, in a cohort of 100 epilepsy patients, 50 might have focal seizures, but only two patients have auditory auras. If all patients with focal seizures had auditory auras, we could almost use the term interchangeably. To make a long story short, assessing meaningful phenotypes has something to do with the information content of the phenotypes. And this is where ontology-based phenotypes might have advantages over a structured dataset.
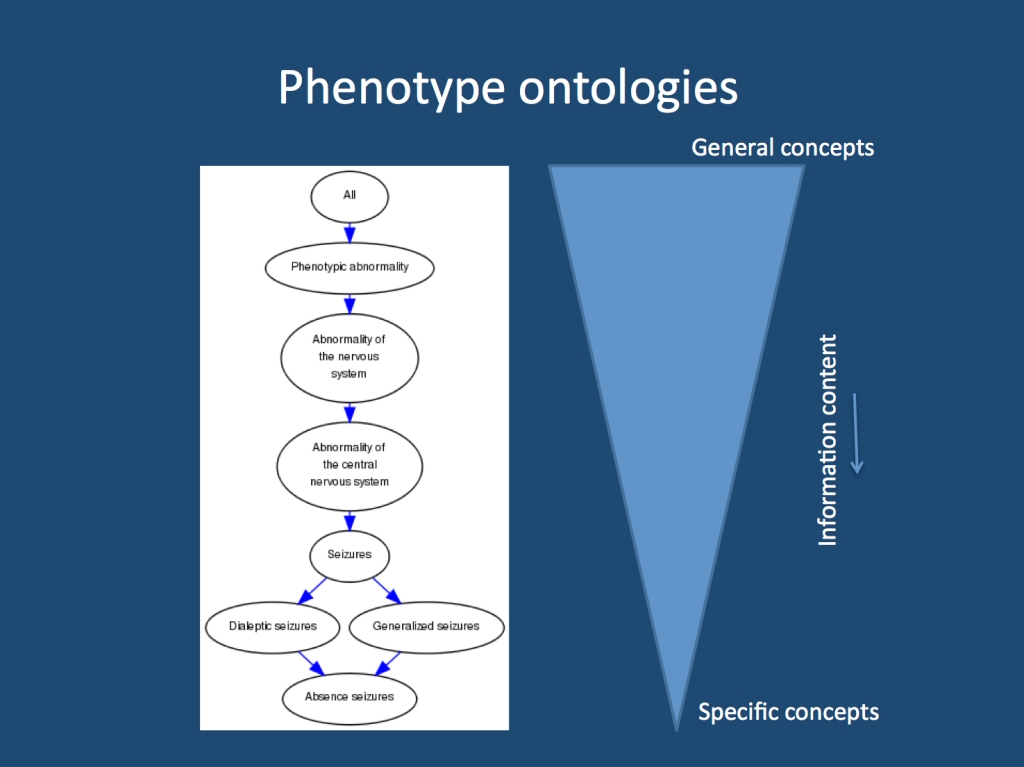
A branch within the ontology tree. Within ontologies, general and specific terms are connected through so-called parent-child relationships. The more specific term is a child of a more general term and the information content increases when moving from general to specific terms. Walking up and down the ontology tree can be used for comparing the proximity of two terms and the similarity of two patients.
What is an ontology? A structured phenotype assessment requires each query field to be completed for each patient. If an entry is not present, it is missing and cannot be used for the analysis. If we only had information on a single patient with auditory auras, we could not use this information in a meaningful way if the information is missing for everybody else. To overcome this problem, ontologies generate connections between phenotypic traits by aligning them on a tree-like structure. For example, auditory auras are connected with partial seizures, they are “a child of” partial seizures, as this is referred to in the language of ontologists. In this system, the information on the single patient with auditory features is not lost, but adds to the information of patients with partial seizures. We have added ~500 epilepsy-specific terms to an already existing framework, the Human Phenotype Ontology (HPO) in preparation for the launch of BENCH-RES.
The age of epilepsy ontologies. Ontologies have one remarkable feature: they are expandable. If we would like to add data from two different centers, which use different levels and classifications for phenotyping, we can merge this data using an ontology tree. This might have advantages in the RES project, where we investigate many epilepsy phenotypes that are complex and difficult to classify. The ontology helps us to add data without requiring specific features to be assessed in all other patients. We think that an ontology-based phenotype structure might be helpful in genetic studies and we have looked at ways to test this hypothesis.
Last common ancestors. Two terms in an ontological tree have a last common ancestor. If we walk up the ontological tree towards the last common ancestor, information gets lost as the more general terms are more frequent in the patient cohort. Therefore, the connection between two terms in a phenotype ontology can be measured by the amount of information that is lost on the way towards the last common ancestor. Likewise, two patients with multiple phenotypic features can be compared by determining the minimal loss of information when all terms are compared with each other. Again, information content can be determined through the frequency of a term in the study cohort, in our case 171 patients. A term that is present in one or two patients has a higher information content (IC = 171/1 = 171) that a term than is present in half of the patients (IC = 171/85 = 2).
A real world example. Using this method, we can define similarity between patients. We looked at the similarity of all patients in BENCH and determined the similarity among them based on information loss of phenotypic traits in the ontology. We then looked at the patients that were most similar to each other. Interestingly, we found five patients with idiopathic West Syndrome, i.e. West Syndrome with good outcome. This group stood out in a patient cohort that is usually characterized by severe epilepsies with poor outcome. We think that this might be a good proof of principle, but it is also a slightly strange finding. We are currently in the process of checking biases and systematic errors that we might have committed. However, as of now, we are happy that we have found a way to compare heterogeneous phenotypes using an unbiased data-driven approach.
A thank you. Entering data into BENCH is a tedious process and I would like to thank all the EuroEPINOMICS partner who helped generate the data that we have now. In particular, I would like to thank Johanna Jähn and Sarah Weckhuysen for the long hours they spent on managing and curating the BENCH database. I would also like to thank Peter Robinson, who has initially developed the Human Phenotype Ontology and has supervised our contribution of epilepsy-specific terms to the HPO.



