The story continues. This week, I am trying to catch up with a number of recent papers in the field of neurogenetics. A recent publication in Nature Genetics highlights the role of de novo mutations identified through exome sequencing in schizophrenia. The authors also look at control data and compare their findings with the growing body of data available for autism research. And while many aspects regarding de novo mutations become more clear with every study published, the real difference is sometimes difficult to grasp. Continue reading
Category Archives: Publications
Genome meets Connectome: gene networks and brain microstructure
Genetic imaging. There are two major fields in epilepsy research – functional imaging and genetics. Both fields live parallel lives and hardly ever interact. When they do, the interaction is usually short-lived and full of disappointments, as nothing has really ever worked. However, a grant application due today has led me to a recent publication in the Journal of Neuroscience, which combines imaging and GWAS. And believe it or not, the ion channels are back. Continue reading
De novo mutations in severe intellectual disability
Diagnostic exome sequencing. Severe intellectual disability (ID) is unexplained in the vast majority of patients and is thought to be genetic. The genetics of intellectual disability has traditionally focused on the X chromosome, where more than 100 possibly causative genes for ID are located. But other, autosomal genes are also found in large number of cases. A recent study in the New England Journal of Medicine now reports on trio exome sequencing in patients with unexplained severe intellectual disability. The authors identify causative de novo events in a large proportion of patients. Interestingly, more than half of their patients had epilepsy. Continue reading
Double Impact
Second hits. Genomic disorders are genetic disorders due to recurrent microdeletions or microduplications, i.e. small losses or gains of genomic material that happen again and again due to existing breakpoints in the human genome. Intriguingly, additional large microdeletions or microduplications can be identified in some patients with genomic disorders. A recent study in the New England Journal of Medicine tries to explain why. Continue reading
To do: read ENCODE papers
ENCODE will change the way we analyse genomes. The comparison of long non-coding RNA and transcription factor binding sites will require more CPU time. Anything else? I don’t know, I am only writing this because Ingo asked me to. It’ll take time to study the 30+ papers, sift through the data and discuss it with colleagues. Only then, something like that understanding we hear so much about can happen and I am sure it will in journal clubs around the globe in the next weeks. But smaller things might already be interesting.
The years of our fathers: paternal age and the rate of de novo mutations
Aging fathers. An increase in risk of aneuploidies, i.e. chromosomal aberrations such as Trisomy 21, is well established with maternal age. Whether the paternal age also increases the risk for disorders in the offspring had long been disputed. However, a connection between paternal age and autism has been found in recent years. Now a recent study in Nature finds a surprisingly strong correlation on the genetic level… Continue reading
Will the relevant SNPs please stand up
The flood of variants. Every re-sequencing of a genome leads to many more variants than can be validated with functional assays. Many strategies exist to select the candidate variants. Filtering on criteria might remove all variants so efforts are focused to re-rank the list of variants such that the most promising appear on top. A recent review in Nature Reviews Genetics wants to give users a hand with using the bioinformatics tools available. As a bioinformatician, I find a number of important points missing.
Somatic mutations affecting the mTOR pathway in hemimegalencephaly
Mutations, but not germline. Many of the genetic alterations that we aim to investigate within the EuroEPINOMICS projects are so-called germline mutations. In the case of de novo events, these mutations have occurred in the germ cells themselves or in very early development. In the case of autosomal dominant or recessive inheritance, the mutations have been transmitted from parents. In either case, the mutation can be found in every cell of the body. Cancer research is mainly focussed on somatic mutations, which give rise to malignant transformation in already differentiated tissues. In fact, many of the techniques that we currently use in neurogenetics were developed to study somatic genetic aberrations. Array comparative genomic hybridization for example, had initially been established for these purposes before expanding the focus to germline microdeletions and microduplications. While the role of somatic mutations in cancer research is well established, the role somatic rather than germline genetic alterations play in other disorders is mainly speculative. Some initial evidence for somatic point mutations has recently been found in Proteus syndrome, a rare overgrowth syndrome. Activating somatic mutations in AKT1 have recently been identified in this disorder. A recent paper by Lee and colleagues now identifies mutations in several genes in the mTOR pathway in patients with hemimegalencephaly, a severe form of brain malformation. Continue reading
One fish, two fish, red fish, blue fish – KCTD13 and neurogenetic studies in zebrafish
Microdeletions in seizure disorders. In a recent paper in Nature, Golzio and colleagues identified KCTD13 as the main driver for the neurodevelopmental phenotype of the 16p11.2 microdeletion. Small losses of chromosomal material as found in microdeletions usually affect several neighbouring genes. Many deletions are due to the particular duplication architecture of the human genome and are canonical, i.e. they always have the same size and include the same genes. The same duplication architecture also makes these variants relatively common, and the full impact of microdeletion-associated genetic morbidity has startled the neurogenetics. The recent five years have led to the identification of several epilepsy-related microdeletions including variants at 15q13.3, 16p13.11 and 15q11.2. There are further microdeletions that are usually found in patients with autism or intellectual disability and to a lesser extent in patients with epilepsy. The 16p11.2 microdeletion, the first microdeletion to be identified through a large-scale association study, is one of these variants.
From deletion to causative genes. For many microdeletions, the statistical evidence for the association with a particular phenotype is often beyond reasonable doubt given that several thousands samples can be included nowadays. The identification of the underlying causative gene, however, is extremely difficult. It is technically challenging and time-consuming to investigate all included genes functionally through conventional model systems. The function of many genes included in microdeletions are not related to ion channels, the best known pathological substrate in epilepsies, and hampers testing effects through established electrophysiological techniques. Finally, microdeletions only lead to hemizygosity, i.e. the second copy of a gene should still be expressed at lower level, requiring model system looking for a quantitative rather than qualitative change. The bottom line is that epilepsy researchers are stuck without suitable model systems, which would allow for a medium-size throughput screening for genes in these deletions. This is where Danio rerio comes into play.
The zebrafish as a model for neurodevelopmental disorders. The zebrafish (Danio rerio) is a good model system for genetic and developmental research. The technologies for genetic manipulation are highly advanced. In addition, embryos are transparent and develop externally. Furthermore, a zebrafish develops quickly and produces a large number of offspring. For her studies on developmental genetics using the zebrafish as a model system, Christiane Nüsslein-Volhard received the Nobel Prize for Medicine in 1995.
Screening of the candidate genes of the 16p11.2 microdeletion. Golzio and coworkers focussed on a peculiar aspect of the 16p11.2 microdeletion as an outcome parameter for their genetic screening – macrocephaly, i.e. an enlarged head circumference. In contrast, patients with the corresponding 16p11.2 microduplication often show microcephaly, i.e. a reduced head circumference. Golzio and colleagues deviced a system to measure head circumference in zebrafish embryos and then overexpressed the 29 genes contained in the 16p11.2 microdeletion in the developing embryo. Strikingly, only KCTD13 resulted in microcephaly. Macrocephaly was seen when KCTD13 was knocked-out with a morpholino. This demonstrated that up- or downregulation of KCTD13 affects head size. The authors went on to show that these differences in head size are driven by differences in neuronal proliferation. KCTD13 is highly expressed in the human forebrain and recent studies have suggested a role for excessive neurons in the frontal lobe in autism.

Figure 1. Study design by Golzio and coworkers to identify KCTD13 as the main gene within the 16p11.2 microdeletion responsible for micro- and macrocephaly. Neuronal proliferation or apoptosis underlies this phenomenon.
Application to epilepsy research. The authors combine a clever screening strategy with a convincing follow-up study, highlighting the potential of zebrafish studies in neurogenetics. However, head circumference is not identical with autism and only represents a surrogate parameter. Therefore, even though the authors emphasize the role of head circumference as an essential part of the 16p11.2 phenotypes, it only represents a minor aspect of it. Nevertheless, the authors demonstrate that Danio rerio is a good model system for medium-throughput screening strategies, and epilepsy models in zebrafish do exist, suggesting that this study design might help decipher the plethora of candidate genes arising from the genetic studies in EuroEPINOMICS.
No use in studying gene-gene and gene-environment effects in complex diseases?
Genome-wide association studies (GWAS) have improved our insight into the genetics of complex diseases but have fallen short of initial expectations, leaving the majority of the heritabililty to be explained. Interactions of genes with the environments and with each other receive a fair share of the blame for the lack of progress despite the widespread efforts. The large number of possible interactions, however, currently still limits progress in this field. A dedicated and growing group of computer scientists and geneticists now study gene-gene effects in the hope of shedding light on complex diseases. Initial results were hopeful, even in the field of epilepsy genetics.
Now, a group of Harvard based biostatisticians presented simulations for breast cancer, type 2 diabetes and rheumatoid arthritis that include gene-gene and gene-environment effects. Their interpretation reads bleak: little predictive power can be gained by including the additional dependencies, which means that all the CPU time consumed currently for their analysis is only warming the planet and the hearts of computer scientists.
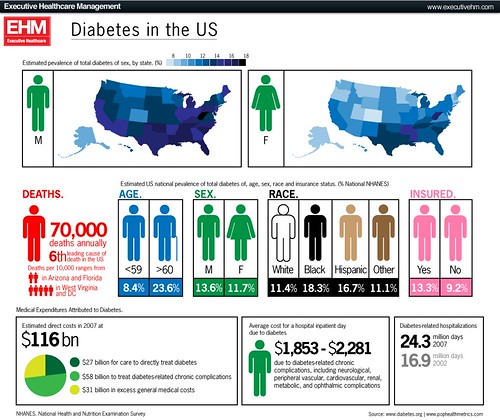
The large number of cases diabetes and many other complex widespread diseases are not explained easily. And the Aschard study suggests that it will remain so for the immediate future despite the progress in sequencing technology.
Negative predictions from experts for their own domain usually receive a negative backlash. The study could probably be attacked on the grounds that the authors selected a large number of parameters, some from probably little more than thin air. But the geneticists on twitter remained silent. Is this acceptance already? Maybe the critics still lie exhausted from attacking Vogelstein’s negative predictions from a couples of months ago.
If the statistical model and parameter choices find widespread acceptance, it would mean that it is virtually impossible to explain many complex diseases from genetics alone to a sufficient degree. As individual studies of the interactions of two SNPs are difficult enough, many cases of complex diseases will remain unexplained. Despite all the efforts, it would be almost as dark as before we had high-throughput sequencing facilities.

