Sequence databases are not the only repositories that see exponential growth. The internet helps companies to collect information in unprecedented orders of magnitude, which has spurned the development of new software solutions. “Big data” is the term that stuck with it and blew life into the data analysis. Widespread coverage ensued, including a series of blog posts published by the New York Times. Data produced by sequencing is big: Current hard drives are too slow for raw data acquisition in modern sequencers and we have to ship the discs because we lack the bandwidth to transmit the data via the internet. But we process them only once and in a couple of years from now they can be reproduced with ease.
Large-scale data collection is once again hailed as the next big thing and spiced with calls for a revolution in science. In 2008, Wired even announced the end of theory. Experimental scientists make good use of hypotheses and targeted experiments under the scientific method the last time I checked though. A TEDMED12 presentation by Atul Butte, bioinformatician at Stanford is symptomatic in it’s revolutionary language and caused concern with Florian Markowetz, bioinformatician at the Cancer Center in Cambdridge, UK (and a Facebook friend of mine). Florian complains and explains that the quantitative changes in the data does not lead to a new quality of science and calls for better theories and model development. He’s right, although the issue of data acquisition and source material had deserved more attention (what can you expect from a mathematician).
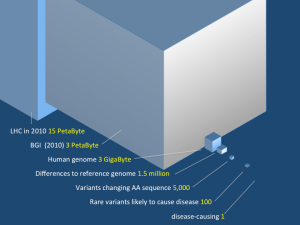
The part of the data we care about in biology is quite moderate but note that the computing resources of the BGI are in league with the Large Hadron Collider.
We don’t know what to expect from e.g. exome sequencing for a particular disease and the only way to find out is to do the experiment, look at the data, come up with guestimates and confirm your finding in the next round. Current data gathering and analysis projects in the life sciences won’t be classified as big data by the next sweep of scientists anyway. They are mere community technology exploration projects using ad hoc solutions.


