Isolates. Last week, the FinnGen biobank went live, and Nature dedicated an entire issue to the launch of this initiative. In brief, FinnGen is a large Finnish research project providing genomic and clinical data from a Finnish biobank with the aim to provide new insights into human disease. Finland is an isolated population, which offers unique insights into the role of rare variants in disease. When I checked the FinnGen database for association with SCN1A, I was surprised that three missense variants have been associated with various diseases. Here is what a founder population can tell us about the various roles of SCN1A in human disease.
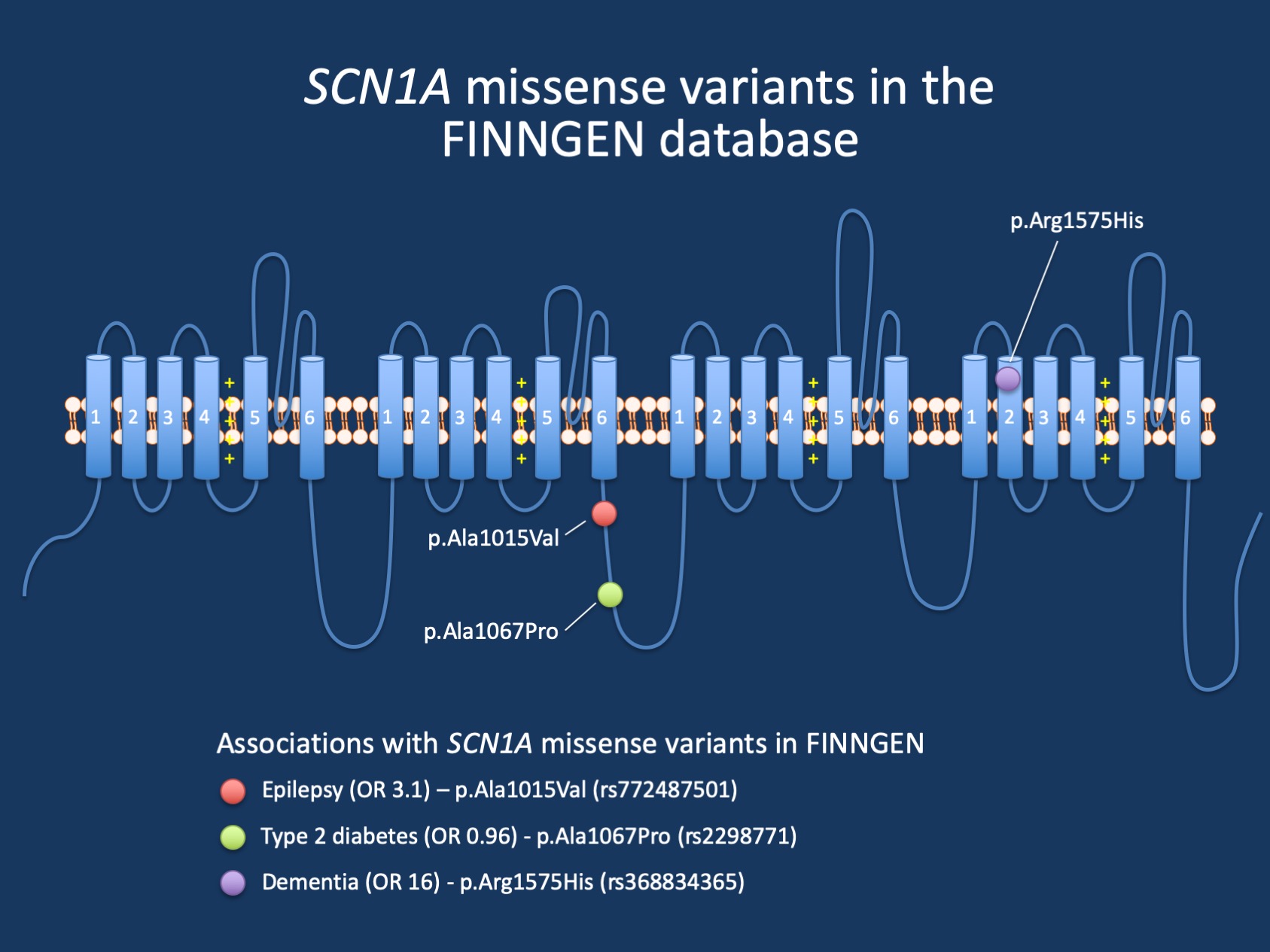
Figure 1. The FinnGen PheWeb database provides an interesting insight into rare missense variants in SCN1A and their associations with human disease. In brief, in addition to the p.Ala1015Val variant, which increases the risk of epilepsy by a factor of three, the p.Arg1575His transmembrane variant increases the risk of dementia by a factor of 15. Conversely, the p.Ala1067Pro variant is slightly protective against type 2 diabetes (https://r8.finngen.fi/gene/SCN1A).
FinnGen. With an entire series of Open Access papers, FinnGen went live last week. In addition to genotyping more than 200,000 individuals, the FinnGen team imputed more than 16 million variants from a reference genome panel specific for the Finnish population of almost 4,000 genomes. This dataset was then combined with 2,800 disease endpoints that the FinnGen team generated from health registries, hospital discharges, and medication prescriptions. Associations with these disease endpoints can be queried through the FinnGen PheWeb browser. This search tools allows for a query based on phenotypes and genotypes. Here is what I found when I searched for associations with SCN1A.
Missense. In my initial search, I focused on SCN1A missense and loss-of-function (LOF) variants. As the dataset does not include novel variants, and all variants had a small minor allele frequency (MAF), I did not find any SCN1A protein-truncating variants (PTV). However, three missense variants associated with different phenotypes caught my eye. Let me state that none of these variants achieved genome-wide significance, but were nominally associated. Here are the three variants that were associated with disease in the FinnGen cohort.
Epilepsy. The strongest association was between the p.Ala1015Val variant and epilepsy with an odds ratio of 3.1. With a MAF of 4.9e-4, this variant is unique – it is a relatively rare population variant that fills the gap between the ultra-rare variants with a strong effect size and common variants with a small effect size. Often, for rare variants, there are hidden population substructures that may create spurious associations. However, the homogeneous population structure of the Finnish population makes it possible for the disease association for this variant to emerge.
Dementia. The association between the p.Arg1575His transmembrane variant and the dementia outcome had the strongest effect size of all three SCN1A variants, with a roughly 16-fold risk. This association is entirely new and may hint at either specific epilepsy phenotypes included in the dementia outcome measure or a new gene-disease association that was unknown before. Currently, it would be pure speculation to attribute any causal mechanism, but this finding clearly warrants further follow-up. This variant is absent from gnomAD, highlighting a dementia risk factor specific to the Finnish population.
Type 2 Diabetes. If the association with dementia was surprising, a protective effect of the p.Ala1067Pro against type 2 diabetes (T2DM) is highly unusual. SCN1A has some expression in lung and the pituitary gland in addition to the well-known expression in brain. Therefore, ascribing any mechanism to this protective effect would be difficult. Given that the protective effect is very small and (to my knowledge) previously undescribed, this is a finding that is hard to reconcile. However, I typically tell our trainees that genomic evidence is usually stronger than any functional consideration. Therefore, if this finding holds true, it might provide a new insight in a more complex role of SCN1A than previously associated.
What you need to know. Large population databases allow for a glimpse into the genomic architecture of human disease, and information on population isolates enables us to assess entire variant classes that are typically difficult to assess: rare (but not ultrarare) variants with a modest effect size. In this blog post, I wanted to provide a first glimpse on how databases such as FinnGen can provide us with new insights into the disease associations of genes that we typically assume to know well. The three SCN1A FinnGen variants represent interesting candidates for functional studies to provide evidence on how missense variants found in the population can increase risk for neurological and non-neurological disorders.


