Area X. Zebra finches are a small bird species that originate from Australia and can be found all around the world. They are highly social birds and even though some zebra finches may sometimes get aggressive when defending their territory, they are generally polite if unprovoked. Seven percent of all zebra finches have interruptions and repetitions in their bird songs which is a naturally occurring variation on how zebra finches communicate. When the same phenomenon occurs in humans, it is referred to this as dysfluency or stuttering. Even though there are many myths around the causes of stuttering, developmental stuttering, the most common form of dysfluency, is a neurodevelopmental disorder with a strong genetic component as shown by twin and family studies. Dysfluency is also a phenomenon that I know extremely well given that I am a person who stutters myself. In 2013, I wrote my first blog post on the genetics of stuttering, telling the story of how my differently wired brain tripped up an epilepsy neuroimaging study. Here is a 2022 update on one of the fascinating conditions that contributes to human neurodiversity.
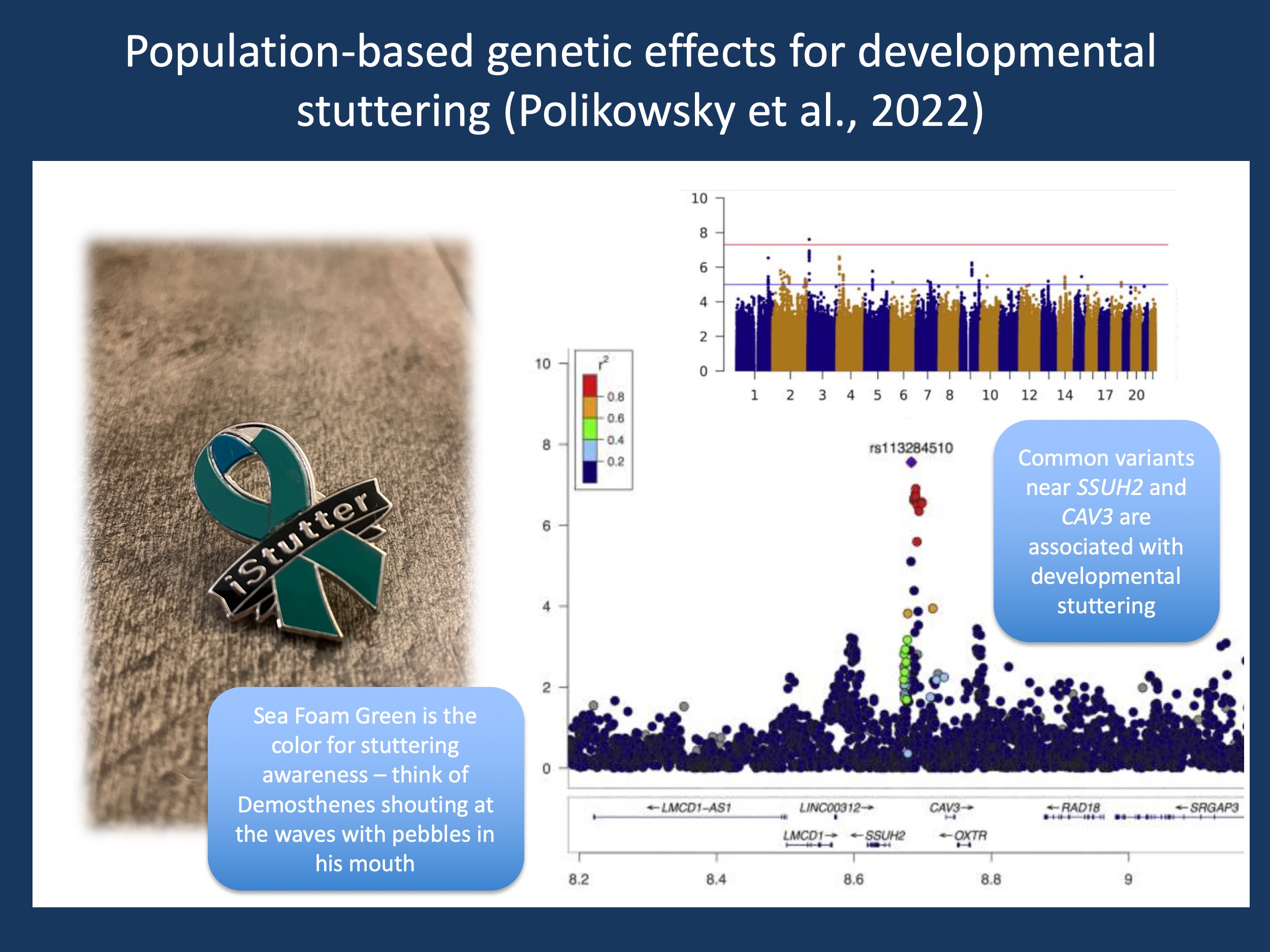
Figure 1. My stuttering awareness pin is a bit battered, having been in daily use since International Stuttering Awareness Day 2021 (10/22/2021). Polikowsky and collaborators published the first genome-wide association study in developmental stuttering, comparing 1,345 clinically ascertained cases, 785 self-reported stuttering cases, and >14,000 population controls. A common variant close to SSUH2 (rs113284510) showed the strongest association, even though the SSUH2 does not have any known connection with the CBGTC loop. Nevertheless, this study highlights the fact that developmental stuttering is a promising target for modern genetic studies with several additional studies on the horizon that are hopefully published soon (edited from https://doi.org/10.1016/j.xhgg.2021.100073 under a Creative Commons license)
The biology of stuttering. I gave the name “Isaac” to the iPad that I bought for telemedicine during the pandemic. Isaac has accompanied me through hundreds of telemedicine encounters and through our telemed research since. It is named after Isaac Newton who stuttered. However, I could have also named it after Charles Darwin, Theodore Roosevelt, King George VI, Winston Churchill, Samuel L. Jackson, James Earl Jones, Emily Blunt, Bruce Willis, Elvis Presley, Marylin Monroe, or Kendrick Lamar. All these people listed here are stutterers. The gender imbalance on this list is no oversight but on purpose. Developmental stuttering typically affects up to 4% of all children, but most children grow out of it. However, men are four times more likely to have persistent developmental stuttering compared to women. Even though much about the neurobiology of stuttering remains a mystery, there has been some remarkable progress over the last decade. It is now established that stuttering occurs due to changes in a brain network referred to as the Cortico-Basal Ganglia-Thalamocortical Loop. This network, the CBGTC loop, is an integrated framework of speech production that is typically more prominent in the left hemisphere. Changes at various points in this “stuttering loop” result in dysfluency. Both the cortex and the basal ganglia within this loop have received recent interest. For example, a recent publication found increased iron deposition in the basal ganglia, putting stuttering in the vicinity of NBIA disorders (neurodegeneration with brain iron accumulation), which is a fascinating and so far unexplored angle. In addition, in our own study by Thompson-Lake and collaborators, we describe a family with an inherited difference of Broca’s area, which failed to follow the typical thinning observed in fluent individuals. But what about actual genes?
Nothing about us… Before I mention stuttering genetics, let me quickly slip into the role of a self-appointed disease advocate, given that stuttering is the one neurodevelopmental condition that I truly own. Since my first blog post in 2013, the stuttering community has become much more vocal, and concepts such as Stuttering Awareness, Stuttering Pride or Stuttering Gain are increasingly discussed in the community (#NoDiversitywithoutDysfluency). People Who Stutter perceive things differently compared to the 99% of the world population that does not stutter. The constant awareness that your thoughts may not automatically translate into spoken words is difficult to explain to a fluent speaker. People Who Stutter are natural born word jugglers who constantly manage their blocks, repetitions, and prolongations and are often able to hide their dysfluencies within typically sounding speech (“passing as fluent”). Where does this leave genetics? There is some perception that gene discoveries in stuttering goes against what stuttering advocacy stands for, reinforcing stigma and “othering”. To me as a child neurologist and neurogenetics researcher, nothing is further from the truth. The genetics of stuttering is far behind, and getting on top of the genetics of human dysfluency is liberating rather than stigmatizing – it will help bust some of the lingering myths in the field and replace them with scientifically founded knowledge. However, getting a handle on genetic factors contributing to stuttering has been complicated.
Stuttering genes are hard to find. Over the past decade, several monogenic causes of human stuttering have been suggested, even though most proposed candidate genes have not been replicated so far. What has emerged from some family studies are gene findings in GNPTAB, GNPTG, NAGPA, or AP4E1. These gene findings highlight an involvement of lysosomal pathways that include the protein products of these genes, raising important hypotheses with regards to some of the disease mechanisms. With increased focus on stuttering genetics, many of these findings may hopefully be replicated soon and then be on more solid footing. But in an era of large biobanks and repositories of thousands of genomic datasets, why make it so complicated? Couldn’t we just look?
Stuttering genomics requires new approaches. It doesn’t work. Many large biobanks, including Vanderbilt’s BioVU and the Penn Medicine BioBank only have a handful of participants with a diagnosis code for stuttering in tens of thousands of recruited research participants. The pediatric sphere is no less bad – in the >13,000 individuals included in the DDD study, not a single individual was phenotyped as having a stutter. People Who Stutter often pass as fluent, but quite honestly, I was unaware of the full extent of this – stuttering is truly EMR invisible and not traceable through the approaches that we can use for common disorders such as hypertension or asthma. In a recent publication from the Vanderbilt biobank, Shaw and collaborators tried to overcome this problem by using a phenome classifier, using a predicted phenotype based on Electronic Medical Record data that was derived via machine learning. The authors identified common variants in CYRIA and ZMAT4 to be associated with stuttering, a first in the field. At the same time, Polikowsky and collaborators published the first Genome-wide Association Study (GWAS) on stuttering that had a variant in the upstream region of SSUH2 (rs113284510) to be associated with stuttering. Without discussing these genes close to the association signals in too much detail, these findings suggest that a critical step has been taken and that genetic studies in stuttering are no longer a thing of the past. However, the difficulties in identifying individuals who stutter from large biobanks and the lack of repeatedly confirmed monogenic etiologies points out that stuttering genetics requires new approaches – it is a neurodevelopmental disorder that will facilitate the development of new concepts and technologies. In a very literal sense, stuttering is teaching the world to listen.
What you need to know. Stuttering genetics has made gigantic leaps forward. Several monogenic causes have been identified in large families, and common genetic risk factors are slowly emerging from creatively designed study paradigms. I am looking forward to the next 5-10 years in stuttering genetics that have the capacity to completely recalibrate our concepts and theories with regards to the causes and contributors to dysfluency.


