The negative exome. Despite writing a lot about the power of next generation sequencing technologies to provide a genetic diagnosis in individuals with severe epilepsies, it is important to remember that most exome tests performed in a diagnostic setting are negative. Even the most optimistic studies do not find a diagnostic yield that exceeds 40%. However, what can be done about the 60-70% of patients who had undergone exome sequencing, the current gold-standard diagnostic testing, but have received a negative test result? A systematic re-analysis after 12-24 months is currently considered one possibility to make sense of existing exome data. In a current publication, the Epilepsy Genetics Initiative (EGI) reports their results of a systematic research-based re-analysis in 166 individuals with epilepsy. In eight individuals, a novel diagnosis could be achieved, including novel genes not known at the time of the initial report and novel mechanisms such as alternative exons. With a diagnostic rate of 6%, this study provides a unique benchmark of what can be expected when exomes initially come back as negative.
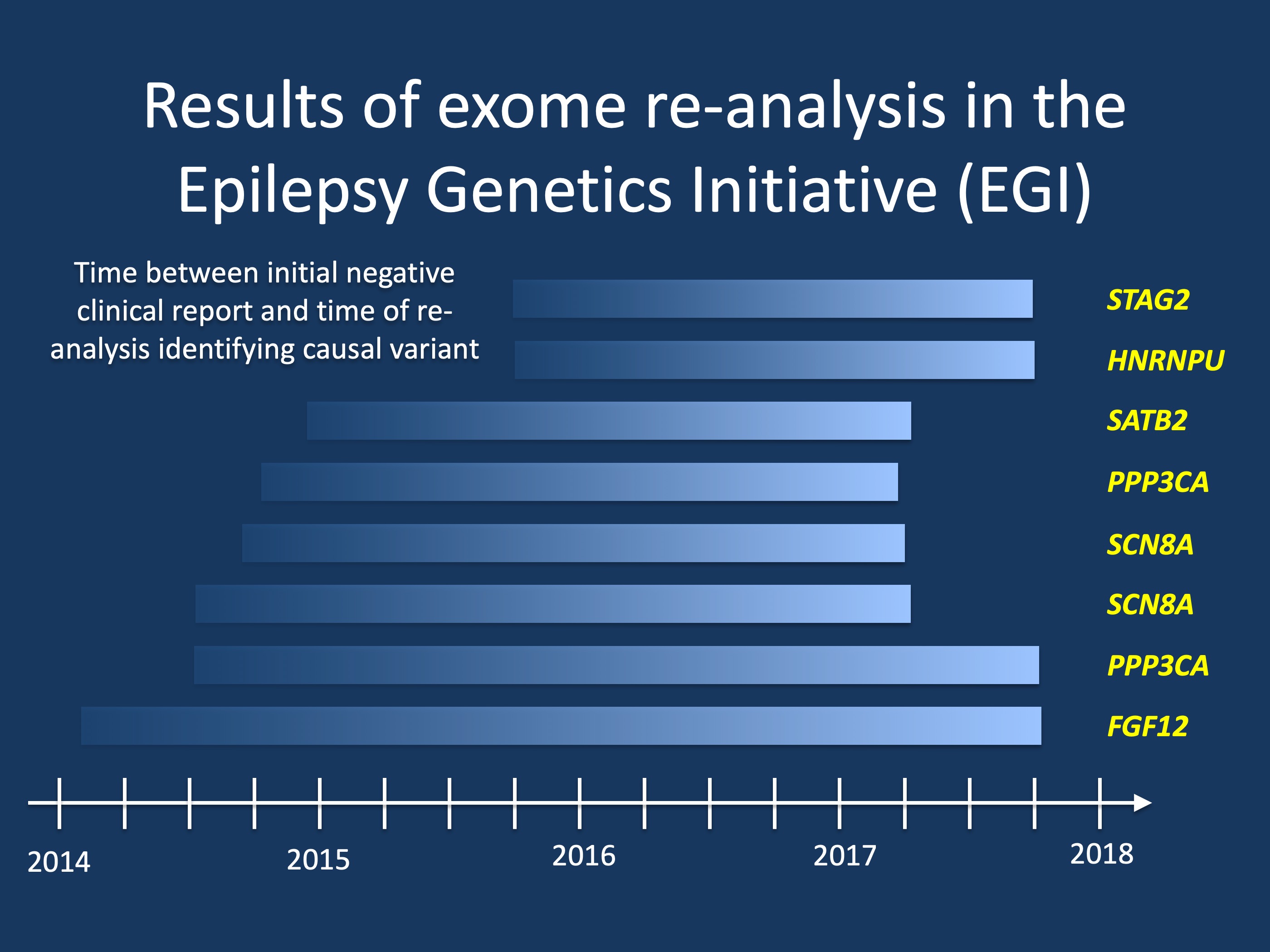
Figure. The Epilepsy Genetics Initiative (EGI) analyzed 166 exome sequences and provided a novel diagnosis in eight individuals, resulting in a diagnostic rate through re-analysis of 6%. The reasons for a gene identified through re-analysis ranged from novel genes unknown at the time of the initial report (STAG2, HNRNPU, SATB2, PPP3CA, FGF12) to novel disease mechanisms such as de novo variants in alternative exons in SCN8A.
The Epilepsy Genetics Initiative. Exome re-analysis is currently a hot topic and is provided by most diagnostic laboratories, either as a complementary service or as a per-fee service. However, there is currently no standard on what constitutes a sufficient re-analysis and some of the information on novel candidate genes may be residing in the research sphere only. Many laboratories currently report novel candidate genes through tools such as GeneMatcher that links submitting researchers or clinicians who submit the same gene. However, all these mechanisms were initially not fully in place yet when EGI initially took off in 2014 as a signature program of Citizens United for Research in Epilepsy (CURE). In fact, at the time of conception, EGI filled a crucial gap in the epilepsy genetics world by providing a unique service to the patient and provider communities by establishing a service that would guarantee a re-analysis of negative exome results. In the current study in Epilepsia, the EGI consortium now reports the results of four years of exome re-analysis.
Why are exomes negative? There is something odd about genetics as a diagnostic test. Just a comparison: when my family doctor sends a CBC, she will be able to tell me whether or not my white count is elevated. It would be very unusual for her to call me up to inform me that the results were uncertain, and that there are currently no other tests that would help her figure out whether the numbers in front of her explain why I am sick or not. It would likewise be strange if she called me up two years later and told me that the CBC that was initially thought to be stone-cold normal is now considered so pathological that it fully explains my disease. This comparison may be unfair, but these are typical issues when interpreting a genetic test in neurodevelopmental disorders: initially negative results may suddenly become positive due to advances in scientific knowledge that now enable the correct interpretation of the causative gene. This unusual feature is largely due to the fact that the interpretation of next generation sequencing-based testing is a snapshot in time, based on the knowledge available to the analysts at the time of testing. Therefore, looking at exome data again after a certain amount of time may reveal different results, emphasizing how important re-analysis has become in neurodevelopmental disorders, particularly for older exomes.
The EGI results. The EGI study entered the field of epilepsy genetics at a critical time point between 2014 and 2018. During this time, exome sequencing was provided by many different labs that applied different criteria with different degrees of stringency. While obvious genetic diagnoses such as a de novo variant in SCN1A could easily be identified by most laboratories, the standards and procedures for reporting candidate genes was less clear. Some laboratories included candidate genes in their reports, others only including known and established genetic etiologies. With increasing application of the ACMG criteria for variant interpretation and deposition of candidate genes into tools such as GeneMatcher, the field has since harmonized. However, for many older exomes, a systematic re-analysis was missing. This service was provided by EGI. Within the EGI study, eight novel diagnoses were made through exome re-analysis, resulting in a 6% diagnostic rate in addition to securing a diagnosis in three individuals where the initial report suggested a candidate gene, but lacked sufficient evidence.
Novel diagnoses. Two genes in the EGI sample were found in more than one individual. Two individuals with de novo variants in PPP3CA were identified, which supported the initial discovery of this gene as a novel cause of developmental and epileptic encephalopathies. PPP3CA encodes the catalytic A subunit of calcineurin, a calcium- and calmodulin-dependent serine/threonine phosphatase, which is important for calcium-dependent dephosphorylation of dynamin-1 after nerve depolarization. Both individuals with de novo PPP3CA variants had drug-resistant infantile spasms. In addition, two individuals with de novo variants in SCN8A were identified, harboring de novo variants in an alternative version of exon 5 (exon 5a) that is not included in the consensus sequence and typically not captured by many annotation tools and therefore missed. One individual with a SCN8A exon 5a de novo variant had West Syndrome and global developmental delay and the other individual had epileptic encephalopathy starting at 3 months. Both genes highlight the main avenues where exome re-analysis is useful: the description of novel genes not known at the time of the initial report (PPP3CA) and advances in analysis technologies that capture variants that were previously missed (SCN8A exon 5a variants). Additionally, diagnoses were made in individual patients with pathogenic variants in FGF12, HNRNPU, SATB2, and STAG2, recently identified genes for neurodevelopmental disorders that were not known at the time of the initial report.
Revoked diagnoses. In addition to making new diagnosis, EGI also removed false diagnoses for two individuals, including a CHD4 variant that was eventually found to be a population variant, and a VHLvariant where a loss of function would only affect a minor isoform, downgrading the variant from likely pathogenic to uncertain significance. Even though “removing” a false positive genetic diagnosis may appear trivial, this is typically a complex process that is often overlooked. In fact, in addition to a missed diagnosis found through re-analysis, there are a significant number of variants that would no longer be considered disease causing in 2019, including entire genes that have lost their status as epilepsy genes such as EFHC1.
What you need to know.The Epilepsy Genetics Initiative has provided an invaluable service to the community by taking in negative exome data and identifying novel diagnoses in 8/166 patients. This provides a unique estimate of what we can expect from exome data that is initially negative, especially older exome tests performed in 2014 and 2015. If negative, exomes need to be re-analyzed, which is likely the highest yield genetic test that can be performed if a patient has negative exome sequencing. Exome re-analysis in individuals with epilepsy may have a yield of at least 6%, suggesting that even negative diagnostic tests have an almost 1:10 chance to result in a definite diagnosis.


