Transfer RNAs. A critical step in translating a cDNA into proteins involves transfer RNAs (tRNAs) that deliver a specific amino acid to the ribosome based on a specific codon in the messenger RNA. Thereby, tRNAs establish the physical link between the mRNA and the amino acid sequence of the growing protein, an essential cellular function. In order for tRNAs to be loaded with the appropriate amino acids, the organism has developed enzymes specific for the individual amino acids that are referred to as tRNA ligases or, more precisely, aminoacyl tRNA synthetases (aaRS). Despite the ubiquitous role of aaRS in the cell, pathogenic variants in genes coding for specific aaRS result primarily in neurological phenotypes, ranging from peripheral neuropathy to early-onset epileptic encephalopathies. In two recent back-to-back publications, bi-allelic pathogenic variants in VARS are described as a novel cause for epileptic encephalopathy, adding VARS to the list of aaRS genes linked to human disease and solving an almost decade-old mystery.
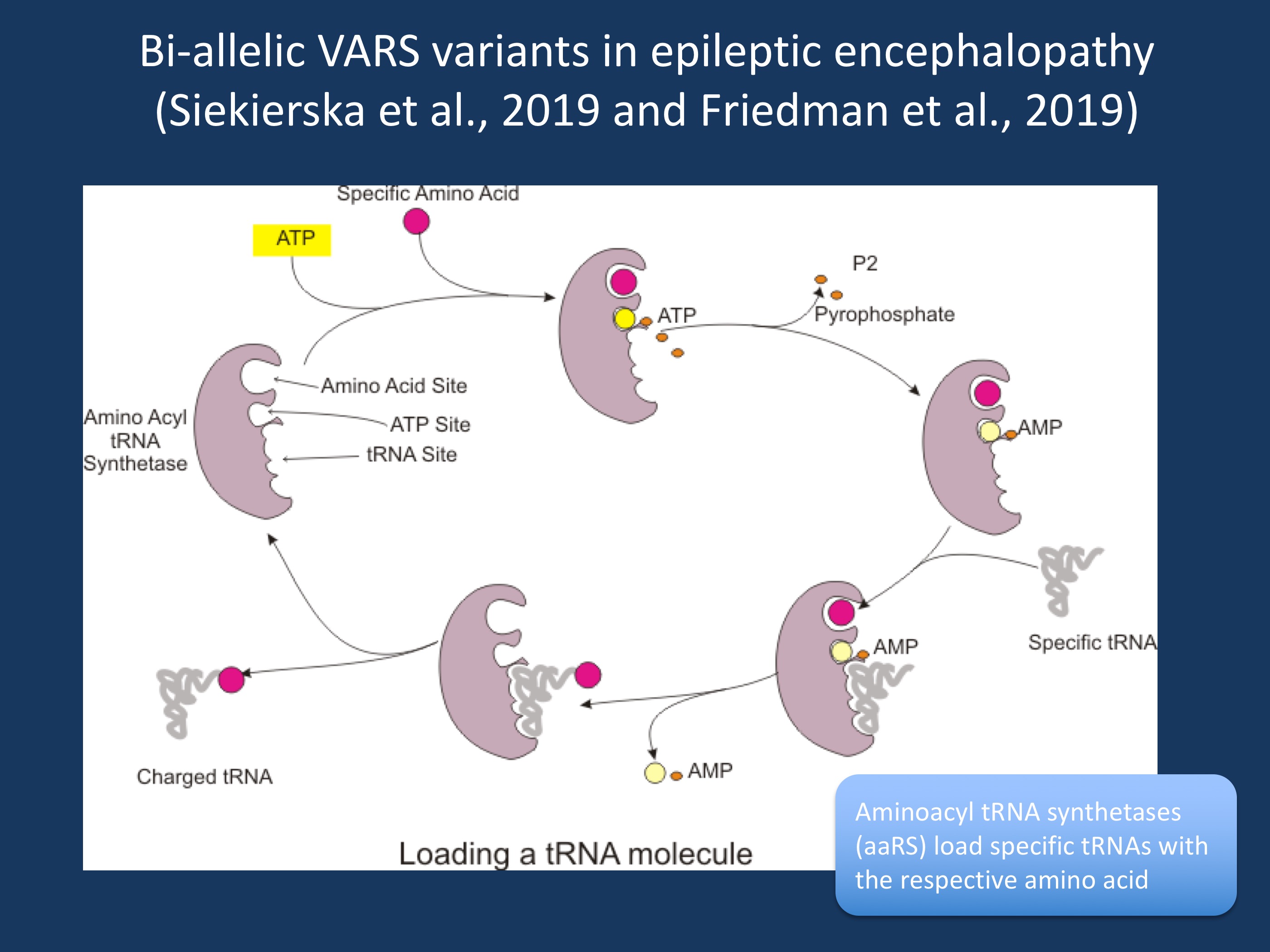
Figure 1. Aminoacyl tRNA synthetases (aaRS) load specific tRNAs with the respective amino acid. As of 2019, more than 30 disorders due to disease-causing variants in aaRS genes have been reported that are involved in mostly neurological phenotypes ranging from peripheral neuropathy to early-onset epileptic encephalopathy. In the publications of Siekierska et al., 2019 and Friedman et al., 2019, bi-allelic VARS variants are reported as a novel cause for early-onset epileptic encephalopathies, often associated with progressive brain atrophy [Image modified under a Creative Commons Attribution-Share Alike 3.0 Unported license from https://commons.wikimedia.org/wiki/File:Charge_tRNA.png)
aaRS. The genes for specific aminoacyl tRNA synthetases are typically named after the single letter code for the specific amino acid, turning the list of existing disease genes within the aaRS gene family into a somewhat similar sounding group of genes: AARS, GARS, HARS, MARS, and YARS are known disease genes for dominant Charcot-Marie-Tooth disease and related peripheral neuropathies; DARS, RARS, KARS, and QARS are genes for severe, recessive neurodevelopmental disorders ranging from hypomyelination with brainstem and spinal cord involvement to epileptic encephalopathies. These are just some examples and as of 2019, 31 aaRS genes have been involved in human disease. It is currently not understood why dysfunction of such an essential cellular mechanism leads to a prominent central or peripheral nervous system phenotype. In two recent back-to-back publications in Nature Communications, Siekierska and collaborators and Friedman and collaborators report a total of 12 families with 18 affected individuals with bi-allelic pathogenic variants in VARS, encoding the aminoacyl tRNA synthetase that loads the amino acid valine onto the respective tRNA.
The VARS phenotype. Bi-allelic pathogenic VARS variants result in prominent global developmental delay with microcephaly and seizures in the majority of patients. The majority of the reported individuals were non-verbal and some individuals had features of progressive cortical atrophy. The VARS phenotype somewhat resembles the disease presentation seen in families with bi-allelic QARS variants and adds to the subgroup of disease linked to aaRS genes that result in early-onset epileptic encephalopathies. We contributed a family to the study by Siekierska and collaborators, solving an almost decade-old mystery of a patient who I had taken care of during my residency and who was included in the early family studies of the EuroEPINOMICS project. Finally, after several years, we were able to pin down the disease gene and demonstrate that the phenotype of VARS encephalopathy can be replicated in a zebrafish model.
The VARS animal model. Within the publication by Siekierska and collaborators, our functional collaborators examined the role of VARS loss-of-function in a zebrafish model, a vertebrate model that can be used for a wide range of screening experiments if the disease model is found to be appropriate. Interestingly, vars knock-out animals had prominent microcephaly due to increase neuronal cell death and reduced swimming activity, a marker of central nervous function in zebrafish. In addition, vars knock-out animals were found to have epileptiform discharges when local field potentials were used. Both sets of experiments complement each other to demonstrate that zebrafish larvae replicate the neurological and epilepsy phenotype seen VARS encephalopathy. These phenotypes could be partially rescued by human wildtype VARS. However, this rescue did not occur when assessing the identified disease-causing variants. Taken together, the VARS phenotype can be assessed in zebrafish larvae, including the inability to rescue the phenotype through human disease-causing variants.
What you need to know. Two parallel publications describe bi-allelic VARS variants as a new cause for human epileptic encephalopathies. VARS joins a growing group of mainly neurological conditions that affect the function of specific aminoacyl tRNA synthetases that load specific amino acids onto the respective tRNAs. By replicating the human disease phenotype in zebrafish, this opens up a way to assess potential mechanisms to overcome the biochemical imbalance due to inefficient valine aaRS activity for future compound screenings in the search for precision medicine directed therapies for these conditions.


