Genome-wide association. While most of the neurogenetics community was focused on exome sequencing and the discovery of novel monogenic forms of epilepsy in the last few years, something quite remarkable had happened in the background. Common variants and genome-wide association studies have made a remarkable comeback. The ILAE Consortium for Complex Epilepsy had slowly worked on increasing sample sizes over time, and the second analysis of common variants in common epilepsies was published in late 2018. Sample sizes have almost doubled since the first study in 2014, and as a result, the number of genome-wide significant loci has tripled. However, the most intriguing finding was something that completely caught me by surprise – more than 30% of the heritability of the genetic generalized epilepsies is explained through common variants, approaching the numbers we see in epileptic encephalopathies explained by monogenic causes. This is one more reason to discuss the recent ILAE GWAS in more detail.
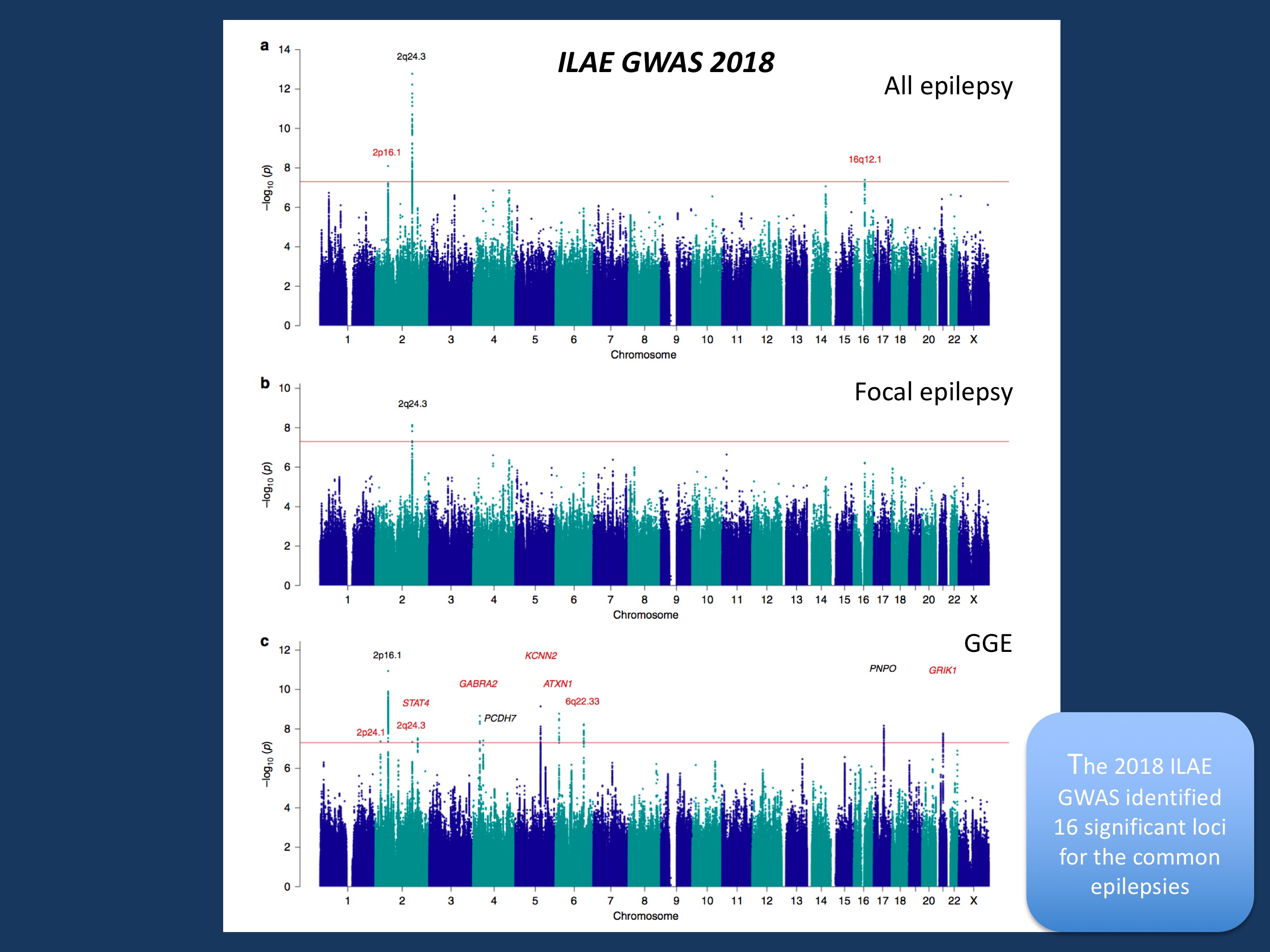
Figure 1 from ILAE GWAS publication. Manhattan plots for epilepsy genome-wide association analyses. Genome-wide association analyses of (a) all epilepsy, (b) focal epilepsy, and (c) genetic generalized epilepsy. Negative log10-transformed P-values (Y-axis) are plotted against chromosomal position (x-axis), the red line represents the genome-wide significance threshold (p < 5 × 10−8). Previously known loci are indicated in black; novel loci in red. The names above each locus represent the prioritized gene in the locus or the name of the locus itself in case of multiple prioritized genes in the locus. Figure and figure legend modified from https://www.nature.com/articles/s41467-018-07524-z under a Creative Commons Attribution 4.0 International License.
GWAS – a brief recap. As we have discussed next-generation sequencing much more frequently than genome-wide association studies (GWAS), here is a brief summary of what these studies do. GWAS looks at common genetic variants, genetic changes in our genome where a significant proportion of the population carries one variant (allele) and the remainder of the population has the other allele. When they affect a single base pair in the human genome, these changes are called Single Nucleotide Polymorphisms (SNPs). SNPs are common, e.g. 30% of the population has an A at a certain position and 70% has a T. Accordingly, SNPs are rarely disease-causing by themselves, as these changes are more common than the frequency of most diseases. However, they can act at risk alleles where they confer a small risk to disease. The state-of-the art method to identify risk alleles is by investigating a large number of SNPs spread throughout the genome in large cohorts and only focus on those alleles that are significantly associated. As several hundred thousand variants are tested simultaneously, a significance level is usually required that is unheard of many other fields of biomedicine, such as p ≤ 5 × 10 e-8. GWAS has been important in the mid-2000s to discovery genetic risk factors for many common diseases such as diabetes or hypertension. In rare neurological disease, next generation sequencing soon became the focus of attention.
The GWAS landscape in neurological disorders. Despite the focus on rare variants, there has been constant progress in assembling larger and larger cohorts for genome-wide association studies, even in neurological or psychiatric disorders. For example, in schizophrenia, the most recent publication includes a staggering 36,989 cases and 113,075 controls. At sample sizes like this, more than 100 risk loci can be discovered. With sizeable GWAS datasets present in most neurological disorders, the overlap between various conditions can be assessed at the level of common variants, allowing for intriguing insight into how different neurological conditions may share common biological pathways. For example, the BRAINSTORM study published in Science compared 25 brain disorders in 265,218 patients and 784,643 controls and found some hint that some forms of epilepsy share some common genetic risk factors with migraine and stroke, but not with multiple sclerosis or Parkinson’s disease.
SNP-based heritability. In addition to finding loci and comparing the overlap between diseases, GWAS at scale can offer something that was thought to be impossible only a few years ago: they can assess how much of the population risk is explained by common variants, the so-called SNP-based heritability. And, quite unexpectedly, the estimates for the SNP-based heritability are both robust and unexpected high. The recent ILAE GWAS shows a SNP-based heritability of 32% in Genetic Generalized Epilepsy (GGE) and 9.2% in focal epilepsies. This is nothing less than breath-taking. A third of the genetic basis of GGE is explained by common variants, taking a giant step towards understanding the cause of one of the most common epilepsy syndromes, roughly catching up with the percentage of individuals with epileptic encephalopathies with monogenic causes. However, this finding comes with two caveats. First, this observation applies to the population in general and is difficult to break down to the individual patient level. A large number of SNPs factor into the heritability analysis, and heritability is an epidemiological term that does not readily apply to individuals. Accordingly, even though the 30% may appear tantalizingly close to our diagnostic yield in epileptic encephalopathies, the actual genetic risk in individuals remains elusive. Secondly, full disclosure, the finding that roughly 30% of GGE can be explained by common variants is not new. It has been around since the initial paper by Speed and collaborators in 2014. One intriguing finding of the BRAINSTORM study was that the SNP-based heritability of GGE is much higher than in other neurological disorders, somewhat setting the GGEs apart of many other neurological disorders.
The 11 novel GWAS loci. Finally, let’s have a look at the individual loci that were found in this second ILAE GWAS that comprises 15,212 cases and 29,677 controls. Overall, 16 loci met the criteria for genome-wide significance. Associated loci are not genes, as most of the significant SNPs are not in located in genes, but next to them. Within the ILAE GWAS, genes hiding behind these loci were prioritized based on their biological link to epilepsy using a scoring system that had also been used in prior studies. This prioritization eventually led to 21 genes. Here is a selection of the suggested risk genes. The most prominent gene groups were both known and novel ion channel genes, most prominently SCN1A. Other ion channel genes were SCN2A, SCN3A, GABRA2, KCNN2, KCNAB1, and GRIK1. This suggests that the monogenic architecture of the epilepsies that includes many ion channel genes is also reflected in the genetic landscape of the more common epilepsies.A specific gene prioritized for Juvenile Myoclonic Epilepsy (JME) was STX1B, a known gene for monogenic Genetic Epilepsy with Febrile Seizures Plus (GEFS+). Finally, the PNPO gene that was already found in the first ILAE GWAS, was found again as a candidate gene in the generalized epilepsies. PNPO deficiency is a severe neonatal metabolic epileptic encephalopathy, and the consistent association with the PNPO locus may suggest a biological connection to the generalized epilepsies. Finally, other than the 2q24 locus that hints at SCN1A, SCN2A, and SCN3A, no further associated loci were found in the focal epilepsies.
What you need to know. The second ILAE GWAS is enormous. This large study compares 15,000 cases and almost 30,000 controls has two major findings. First, 16 loci were identified and may hint at promising candidate genes such as known and novel ion channel genes. Secondly, this study reinforces the role of common variants in the generalized epilepsy, with more than 30% of the population risk explained by common variants. This study helps us understand the causes of common epilepsies and identify further ways how genomic information can help us identify novel biological pathways underlying the common epilepsies.


