Common variants. In addition to the gradual increase in gene discovery due to exome sequencing, there is a field of human genetics developing in parallel that we have not paid much attention to recently. The role of common genetic variants or Single Nucleotide Polymorphisms (SNP) was initially limited to genome-wide association studies, looking at single variants individually. However, more recently, common variants have been assessed jointly in various diseases, resulting in so-called polygenic scores. In a recent publication in Nature, the polygenic contribution to neurodevelopmental disorders is evaluated. Interestingly, there seems to be a very robust contribution of common variants in neurodevelopmental disorders, even in patients with known de novo variants. Here is a brief discussion on why common variants start getting interesting for the neurogenetics field again.
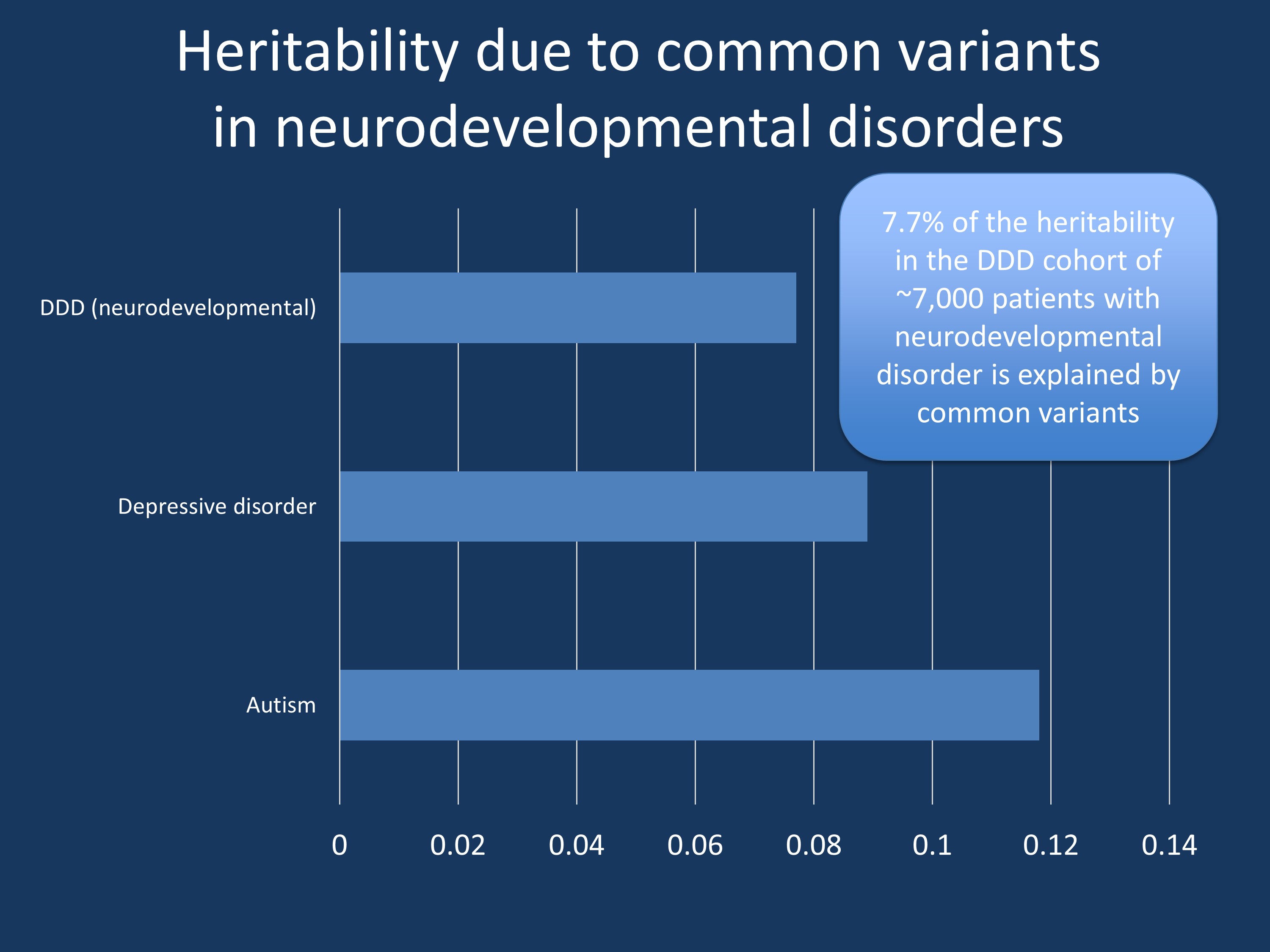
Figure 1. Current estimates for the contribution of common genetic variants to the heritability of various neurodevelopmental and neuropsychiatric diseases. The publication by Niemi and collaborators identified a common variant heritability of 7.7% in patients with a broad range of neurodevelopmental disorders from the DDD cohort.
Polygenic risk scores. Admittedly, I have not paid much attention to what has happened in the field of common variants recently until the recent publication by Niemi and collaborators woke me up. For the last four years, I felt safely reassured that studies focused on common genetic variants with small effect would not mean that much in the neurogenetics sphere. Yes, there were single variant associations that shed some light on underlying pathways, but I never got the impression that the contribution was significant for anything other than hinting at pathways and elucidating the underlying biology. However, this assumption was far from correct. With the massive amount of genetic data that has accumulated over the years, scientists involved in genome-wide association studies have taken the next step by asking the question whether the cumulative effect of common variants has a significant contribution. Variants that are significant in association studies can be combined to a so-called polygenic risk score, the first real-world example on how complex genetics may happen – hundreds, if not thousands of common variants may interact. Polygenic risk scores are increasingly used and help explain a significant population burden of a given disease, with current estimates of up to 10-20% in schizophrenia and other neuropsychiatric diseases.
Common variant risk in severe neurodevelopmental disorders. The current study by Niemi and collaborators, however, takes yet another step and enters the arena of diseases that are usually thought to be monogenic – the severe neurodevelopmental disorders in the DDD study that often have an involvement of several organ systems. The authors first performed a genome-wide association study in almost 7,000 patients and 9,000 controls to assess whether individual variants are associated with the risk for neurodevelopmental disorders. In this analysis, no single variant was associated. However, the authors noted that the overall distribution of p-values was somewhat off. Statisticians use the word “inflated” to describe if a distribution of p-values is showing more significant values that would be expected. This can usually be due to two factors: an artifact due to population stratification or a polygenic “load” of a joint risk mediated by common variants. The authors could demonstrate that this inflation is due to the latter, a polygenic load in severe neurodevelopmental disorders that was subsequently confirmed in a smaller confirmation cohort. This polygenic load explains almost 8% or the overall population risk of severe neurodevelopmental disorders.
Links between common variant scores. The authors then tested whether the polygenic variant signal overlapped with other polygenic signatures that had previously been described. They found that their polygenic score was correlated with known polygenic scores for schizophrenia, lower educational attainment, lower IQ scores, ADHD, and major depressive disorder. However, there was no correlation with scores for Parkinson’s disease, type 2 diabetes, Alzheimer’s disease, or autism. Interestingly, there was no difference in the risk score in patients with and without explanatory monogenic causes. So, what does this mean?
Modifier genes. The study by Niemi and collaborators elegantly demonstrates that de novo variants in severe neurodevelopmental disorders do not occur in isolation. A common variant signal may modify the severity of the phenotypic presentation due to a de novo variant. This means that de novovariants are not black-or-white events, but that the degree of phenotypic severity may be modified by other genetic variants. For example, in patients with recurrent de novovariants in STXBP1 or SCN2A, we often see a range of phenotypic presentations. This variability may be due to the cumulative effect of common variants in the same way that these variants underlie the spectrum of education attainment and intelligence in the general population.
What you need to know. The study by Niemi and collaborators assessed the contribution of common genetic variants in patients with severe neurodevelopmental disorders. The authors find that almost 8% of the population risk is explained by common variants, independent of whether the patient has an explanatory variant or not. This suggests that common variants may modify the presentation of neurodevelopmental disorders.


