Reverse. With the increasing amount of genetic information available in patients with various neurodevelopmental syndromes, some genes will be observed more than once in patients. In a recent study in the Journal of Medical Genetics, the authors trace back the phenotypes of individuals carrying de novo mutations in PURA. However, there seems to be a wide range of clinical features with a seemingly inverse genotype-phenotype correlation.
Study by Hunt and collaborators. Using a large-scale studies on developmental disorders in the UK, Hunt and collaborators identified three patients with de novo mutations in PURA. A fourth patient was identified through a referring clinic in Australia. Patients 1-3 had developmental delay including issues in the neonatal period in some patients. The extent of the delay in patients was highly variable. For example, patient 1 was able to sit independently at the age of 4 years, but was non-verbal and was not able to walk. The delay in patient 2 was milder. She spoke first words at 2 years and started walking at the same time. At the age of 14, she had obvious intellectual disability, but was able to communicate in full sentences. She had some dysmorphic features and was microcephalic. Microcephaly, however, was not observed in other patients with PURA mutations.
In-frame deletion. One patient in the case series by Hunt and collaborators had infantile spasms. Interestingly, this patient had an in-frame deletion, a deletion of three bases, which cuts out a single amino acid while leaving the overall sequencing of the coding frame intact. The in-frame deletion affected one of the conserved regions of the PURA protein, suggesting that this deletion was contributory to the phenotype. In-frame deletions are often filtered out when assessing exomes, as they are often thought of as benign variants. Even de novo in-frame deletions are often suggestive of non-contributory random genomic variation. Therefore, the fourth patient reported by Hunt and collaborators presenting with a de novo PURA in-frame deletion and infantile spasms is a good reminder that we should revisit seemingly benign variation if we have a high clinical suspicion for a causative gene.
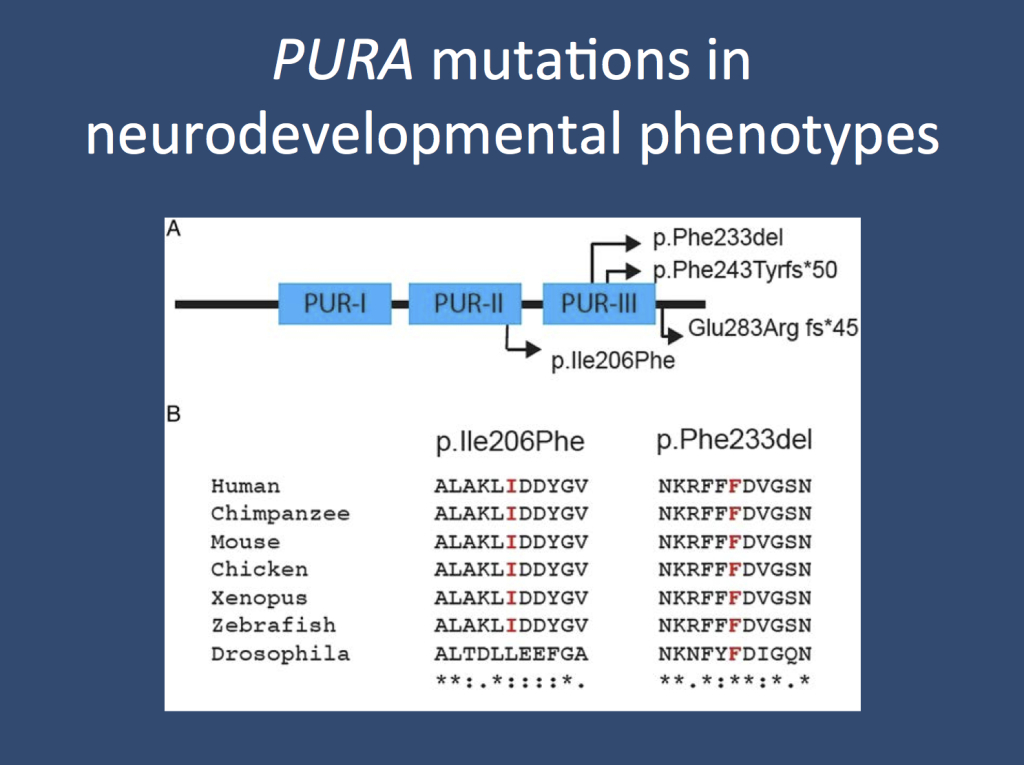
Figures 1a and 1b from the publication by Hunt and collaborators. (a) The mutations in PURA are clustered in the second and third PUR domain, the functional elements of this protein important for DNA binding. PURA binds to single-stranded DNA and is involved in transcriptional regulation. (b) Two of the PURA mutations are highly conserved between species, indicating that they have an important functional role (modified under a Creative Commons license from the publication by Hunt and collaborators)
PURA function. What does the PURA protein do? The PURA protein family is a conserved family of proteins that binds to single-stranded DNA. By binding only to specific sequences, PURA regulates transcription and is also involved in transporting mRNA into neuronal dendrites. The essential function in the CNS is difficult to understand based on this function. However, the important role of PURA in the CNS becomes obvious when PURA-deficient mice are examined. If PURA is missing, mice have various neurological deficits and epilepsy and die prematurely. This suggests that functioning of neurons is deeply dependent on proper and sufficient PURA proteins. Interestingly, PURA-deficient mice initially have no remarkable phenotype and then develop symptoms at 2 weeks of age, suggesting an increasing need for PURA in the maturing CNS.
This is what you need to know. Hunt and collaborators report on four patients with various neurodevelopmental issues and de novo mutations in PURA. The four reported patients have phenotypes ranging from otherwise non-syndromic intellectual disability to epileptic encephalopathy and delays already obvious in the neonatal period. Of interest, the patient with the most severe phenotype appears to have the least suspicious genetic variant, suggesting that the genotype-phenotype correlation is far from clear. The investigation of further cases will show whether there is a common clinical picture for PURA encephalopathy or whether mutations only predispose to a wide range of neurodevelopmental issues.



