What has become of that simple 21,000-gene genome of ours? Today even the definition of gene is no longer clear. What biotypes belong to lncRNAs and what’s the job of unitary pseudogenes? For geneticists dog-paddling in complex diseases another surprise came last year with the announcement that roughly 80% of the genome has some sort of function. Confused? Grab this issue of Genome Research and read the review by Mudge and colleagues, who discuss many examples of the transcriptional complexity within the human genome.
Transcription, splicing, translation. This simple sketch of gene transcription has been decorated by alternative splicing, exon skipping, retained introns and even more bizarre concepts which will run a cold shiver down a geneticists back connecting the complexity to their potential in causing clinical syndromes. And it gets worse: read-through transcripts include exons from two neighboring protein-coding loci, alternative splicing can be highly tissue specific and there is an average of 4 transcripts per protein coding gene. You are aware of bidirectional promoters already, I presume?
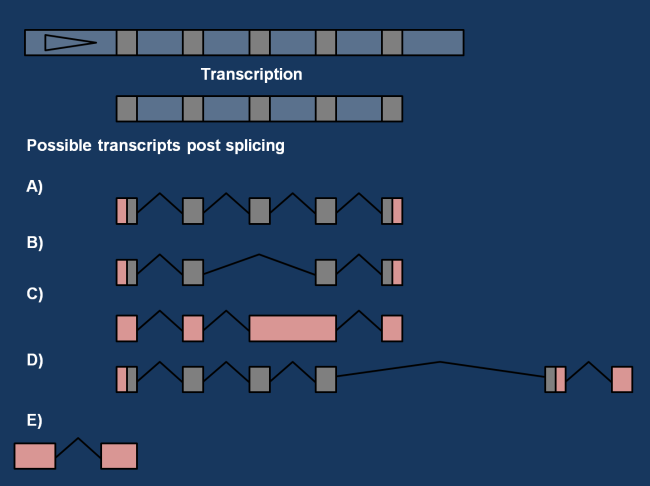 Possible transcripts depicted by Mudge et al., 2013. A) represents the expected classical transcript, whereas B-E) represent possible alternative transcripts.
Possible transcripts depicted by Mudge et al., 2013. A) represents the expected classical transcript, whereas B-E) represent possible alternative transcripts.
What is a functional transcript? The last decade has shown that protein-coding transcripts are not the only biotypes of the genome. GENCODE annotates several other locus biotypes like lincRNAs, antisense lncRNAs, sense intronic IncRNAs, small noncoding RNAs, unprocessed pseudogenes, processed pseudogenes as well as unitary pseudogenes. Mudge and collaborators give several examples how these biotypes can act functional and thereby discuss the concept and designation of functional transcription itself.
Implications in disease research. Whole genome sequencing (WGS) allows for the quick identification of all variation in the individual human genome. The costs for WGS are dropping rapidly and it will become a standard tool in research and diagnostics. If we consider the current international efforts to jointly analyze large patient and control cohorts at a meta-level, it is also likely that we will identify most disease associated variants with modest to large effect within the next years. But what happens when complex diseases turn out to be really complex genetic? How do we crack true complex diseases where the phenotype is not expressed unless more than “XY” mutations across all biotypes are mutated? Current in vitro studies on functional characterization of mutations might be biased due to the fact that analyzed transcript does not reflect the major transcripts in vivo. Can we trust expression array studies, where the probes feature short oligos? We don’t have to consider epigenetics and somatic mutations to be scared. In the last years the major focus was data generation and this trend is likely to continue. But the obstacle has changed from variant analysis to variant annotation, eventually leading to integrative analysis. Mudge and collaborators propose that “the true overlap between transcriptional complexity and functionality will not be gained in the short term”. I don’t know if our high-throughput research has to drop a gear and we will become bench scientists again but at least we can be confident that there will be more than enough to explore and work on.


Pingback: Chutzpah, Zen and Fahrvernügen – the 2013 Sde Boker workshop | Beyond the Ion Channel
Pingback: 9 things you didn’t know about bioinformatics | Beyond the Ion Channel