Genome vs. exome sequencing. Can non-coding regions be skipped in the search for disease-causing variants? Is it worth to pay a higher price for sequencing the whole genome?
The sequencing company Complete Genomics (CGI) is already sounding the death knell for exome sequencing, arguing that the protein-coding genes cover only ~1% of the genome, while many loci identified by GWAS lie in the non-coding regions. CGI maintains that the price difference between whole-genome (WGS) and exome sequencing (ES) has become “less of an issue”. With declining sequencing prices, this will certainly be the case in the future – however, when multiplying the current added costs for WGS with the large numbers of cases and controls required for finding new hits in complex diseases, the proponents of ES have strong arguments. Will WGS explain more than the 10% expected for exome sequencing?
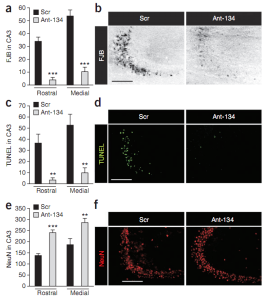
In vivo studies showing how silencing of miR-134 protects against status epilepticus. Graphs and photomicrographs from the dorsal hippocampus of mice 24 h after status epilepticus, treated beforehand with Scr or Ant-134. Counts for three markers are shown, a marker for neuronal death (FJB; a,b), a marker for DNA fragmentation (TUNEL; c,d) and the neuronal marker NeuN (e,f). Mice injected with Ant-134 showed a clear reduction in FJB and TUNEL staining and less of a loss of NeuN staining after status epilepticus, highlighting the neuroprotective effects of Ant-134. Adapted by permission from Macmillan Publishers Ltd: Nature Medicine (E. M. Jimenez-Mateos et al., Vol. 18, No. 7, p. 1087-1094, 2012), copyright (2012)
News from the gene desert. A new study on temporal lobe epilepsy published in Nature Medicine shows that sequencing the non-protein-coding regions can in fact detect major disease-associated changes. The authors investigated the role of a brain-specific miRNA, miR-134, that had previously been implicated in the regulation of neuronal microstructure and found up-regulated in injurious seizures (status epilepticus) and in human epilepsy. When silencing miR-134 expression in mouse models of epilepsy after induction of status epilepticus, the subsequent occurrence of spontaneous seizures and typical pathological features of temporal lobe epilepsy were drastically reduced. Whether the seizure-suppressant actions of miR-134 silencing are anticonvulsant or truly antiepileptogenic effects still needs to be determined. In both cases the molecule used to silence miR-134, a chemically engineered oligonucleotide known as antagomir, could become the basis for a novel therapeutic approach to treat epilepsy.
Implications. The finding lends further support to CGIs statement that disease-relevant genomic loci can be found in gene deserts. There are financial and biological justifications for relying on the ES approach including its proven track record and established miRNAs can be included in the exome capturing array. The sequences of non-coding regions obtained with WGS might turn out to be important but we will need tools to filter variants that are probably causative. Polyphen or SNAP will be of little help for non-coding DNAs.

